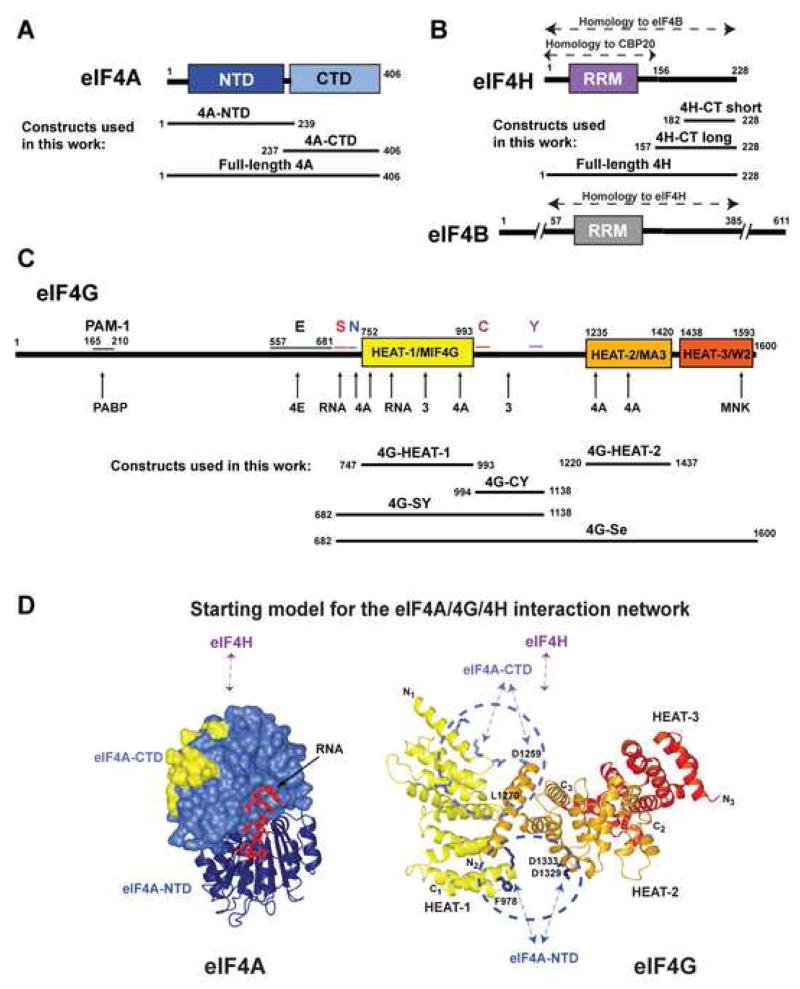
Figure 1. Domain organization of eIF4G, eIF4A and eIF4H.
(A-C) Domain organization of eIF4A (A), eIF4H and eIF4B (B), and eIF4G (C) and constructs used in this work. In panel C, conserved sequence segments in eIF4G are marked above the diagram. Sites of interaction with other proteins and RNA are marked with arrows below the diagram. Abbreviations: NTD, N-terminal domain; CTD, C-terminal domain; RRM, RNA recognition motif domain; 4H-CT-long and 4H-CT-short, long and short C-terminal fragments of eIF4H; PAM-1, PABP-binding motif-1; E, predicted extended eIF4E-binding region; S, RNA-binding region important for scanning, aa. 682–721 (Prevot et al., 2003); N, H1-NT motif, aa. 722–741; C, H1-CT motif, aa. 994–1027; Y, y4G-CT motif, aa. 1118–1136 (Marintchev and Wagner, 2005). (D) Starting model for the eIF4A/4G/4H interaction network. eIF4A-CTD is shown in surface representation and the eIF4G HEAT-1 contact surface (Oberer et al., 2005) is yellow. The interdomain orientation between eIF4A-CTD and eIF4A-NTD (navy ribbon), and the RNA (red wire) are modeled based on the structure of eIF4A3 (Andersen et al., 2006). The interdomain orientation of the HEAT domains of eIF4G (Marintchev and Wagner, 2005) is modeled based on the interdomain orientation in CBP80 (Mazza et al., 2002). The HEAT domains are color-coded as in panel C. Sites of mutations in eIF4G reported to affect eIF4A binding are shown as light blue (top cluster) and navy (bottom cluster) wires. Residues relevant for this work are labeled. The predicted binding sites of eIF4H, eIF4A-NTD and eIF4A-CTD are shown with arrows.