Abstract
Glucocorticoids suppress mast cell activation by inhibiting signaling events as well as transcription of cytokine genes. The inhibition of signaling events has been attributed to upregulation of inhibitory regulators such as Src-like adaptor protein1 (SLAP), downstream of tyrosine kinase1 (Dok1), and dual specificity protein phospahatase1 (DUSP1). As reported here, the upregulation of SLAP and Dok1, but not DUSP1, in the RBL-2H3 mast cell line was inhibited by actinomycin D and was thus dependent on gene transcription. Examination of the gene sequences revealed a glucocorticoid response element (GRE) and a half GRE as potential regulators of the SLAP and Dok1, respectively. As indicated by luciferase reporter assays, SLAP GRE, but not the Dok1 half GRE, robustly activated gene transcription after treatment of cells with glucocorticoids. Binding of the glucocorticoid receptor to the SLAP GRE was verified by chromatin immunoprecipitation assay. These findings further support the notion that the immunosuppressive actions of glucocorticoids are exerted in part through upregulation of inhibitory regulators by various mechanisms. In the case of SLAP specifically, this requires activation of gene transcription through the interaction of the glucocorticoid receptor with GRE.
Keywords: Mast cells, SLAP, Gene transcription, Glucocorticoids, Glucocorticoid response element
1. Introduction
Mast cells play a key role in IgE-dependent inflammatory allergic diseases such as asthma, contact dermatitis, and anaphylaxis as well in autoimmune diseases such as rheumatoid arthritis and colitis (Sayed et al., 2008). Mast cells can mediate both immediate and sustained inflammatory responses through release of histamine and production of inflammatory eicosanoids and cytokines. These responses may be further amplified through recruitment of other inflammatory cells (Gilfillan and Tkaczyk, 2006).
Stimulation of mast cells via its IgE receptor, FcεRI, results in the activation of Src and Syk tyrosine kinases which in turn phosphorylate FcεR1 subunits and the transmembrane adaptor proteins LAT and NTAL (Rivera and Gilfillan, 2006). These phosphorylations allow recruitment of additional signaling molecules and further propagation of signals that include the activation of phospholipase C, calcium mobilization, protein kinase C, phosphatidylinositol 3-kinase, and MAP kinases. These signaling events lead to degranulation for release preformed inflammatory mediators, activation of phospholipase A2 for production of arachidonic acid, and the activation of transcription factors for the generation of various cytokines and chemokines.
The glucocorticoids remain one of the most effective therapies for treatment of mast cell-related disorders. Glucocorticoids act in a time-dependent manner to potently suppress degranulation and cytokine production in mast cells in vitro (Daeron et al., 1982; Berenstein et al., 1987; Andrade et al., 2004; Nakamura et al., 2005). In addition to suppression of cytokine gene transcription (Nakamura et al., 2005), glucocorticoids also inhibit the signaling processes in mast cells (Andrade et al., 2004) by upregulating several inhibitory regulators of signaling by mechanisms that remain undefined. These regulators include downstream of tyrosine kinase1 (Dok1) which activates the Ras GTPase activating protein and thus suppresses activation of the Ras/Raf1/Erk/phospholipase A2 pathway (Hiragun et al., 2005). Another is the dual-specificity protein phosphatase 1 (DUSP1), also known as MAP kinase phosphatase-1. DUSP1 dephosphorylates, and thus inactivates, Erk and p38 MAP kinase in a variety of cell types (Lang et al., 2006) including mast cells (Kassel et al., 2001; Jeong et al., 2003). A third is Src-like adaptor protein (SLAP), which we shall refer to as SLAP1 to distinguished from its recently described homolog SLAP2 (Pandey et al., 2002; Loreto et al., 2002). SLAP1 is known to negatively regulate T and B cell signaling processes (Sosinowski et al., 2000; Tang et al., 1999; Dragone et al., 2006; Myers et al., 2006) and expression of T and B cell receptors in a c-Cbl-dependent manner (Dragone et al., 2006; Myers et al., 2006). As reported recently by our laboratory, SLAP1 is upregulated by dexamethasone in mast cells resulting in suppression of FcεRI-mediated signaling events such as the phosphorylation of Syk, LAT, PLCγ2, and ERK. Release of inflammatory mediators is also suppressed (Hiragun et al., 2006).
The mechanism of action of glucocorticoids at the cellular level is complex and is still a topic of investigation (Clark, 2007). The simple model is that glucocorticoids suppress cytokine gene transcription through the interaction of the ligand activated glucocorticoid receptor (GR) with transcription factors and co-activators to suppress cytokine gene transcription by a process referred to as transrepression. This is in contrast to the activation of gene transcription through the glucocorticoid response element (GRE), a process referred to as transactivation (Ito et al., 2006). Here GR binds either as a dimer to the palindromic response element (GRE), namely GGTACAnnnTGTTCT and variants thereof (So et al., 2008), or as a monomer to sites known as half GRE which consist of the initial or latter part of this sequence (Schoneveld et al., 2004). However, transcriptional regulation of some genes by glucocorticoids requires co-ordinate action of GR with other proteins and transcription factors (Schoneveld et al., 2004). In addition to the negative or positive regulation of gene transcription, glucocorticoids can influence transcript stability by promoting mRNA degradation through upregulation of tristetraprolin, an anti-inflammatory and mRNA-destabilizing agent that recognizes mRNA AU-rich elements (AREs) (Smoak and Cidlowski, 2006; Ing, 2005; Stahn et al., 2007; Stellato, 2004; Clark, 2007). Also, glucocorticoids may exhibit rapid effects that are characterized as nongenomic (Stahn et al., 2007; Stellato, 2004).
We have initiated studies to define the mechanisms for upregulation of the glucocorticoid-inducible inhibitory regulators because these mechanisms are unclear. As reported here, preliminary studies were conducted in the RBL-2H3 mast cell line with inhibitors of transcription and translation namely, actinomycin D and cycloheximide. A search of data banks by the MULAN program indicated potential regulation of the SLAP1 and Dok1 genes by a full GRE and a half GRE, respectively. The DUSP1 gene lacked such elements. The roles of these GREs were investigated by use of luciferase reporter and chromatin immunoprecipitation (ChIP) assays. Our studies revealed that of the three glucocorticoid-inducible regulators examined only SLAP1 was dependent exclusively on gene transcription via GRE for its upregulation.
2. Material and Methods
2.1. Reagents
Reagents were from the following sources: Cycloheximide and actinomycin D from Sigma-Aldrich; polyclonal Abs against GR (P-20) and rabbit IgG from Santa Cruz Biotechnology; protein A-coupled dynabeads, Trizol, and Pfx DNA polymerase from Invitrogen; synthetic DNAs from Integrated DNA technology; reagents for cell culture from GibcoBRL; restriction endonucleases and T4 DNA ligase from New England Biolabs; Pfu DNA polymerase, PCR-based mutagenesis kit, and competent E. coli cells from Stratagene; reverse RNA polymerase and PCR purification kit from Qiagen; dual-luciferase reporter assay system, random primer for cDNA synthesis, genomic DNA purification kit, and dNTP mixture from Promega. Cell transfections were performed with the Amaxa nucleofector kit R.
2.2. Cell culture and experimental conditions
RBL-2H3 cells, were grown in MEM supplemented with 15% fetal bovine serum, glutamine, and antibiotics as describe previously (Andrade et al., 2004). For studies of mRNA expression, cells were plated in 6-well plates (105 cells/2 ml/well) in growth medium for examination of the effects of 100 nM dexamethasone, 100 ng/ml actinomycin D, and 1 μg/ml cycloheximide. The latter drugs were added 20 min before addition of dexamethasone. When ethanol was used as vehicle the final dilution in culture medium was >0.1% ethanol.
2.3. Measurement of mRNA expression
Total RNA was purified from cells of which 1 μg was used for synthesis of cDNA according to the manufacturers’ protocols (Invitrogen and Qiagen, respectively). cDNA was cleared with a PCR purification kit and cDNAs for SLAP1, DUSP1, DOK1, and GAPDH were amplified with the following primers: Dok1 TCT TTC AGG CAG TTG AGG CTG CTA and GCA CGT GCC CAT ACA AAT CCC AAT; DUSP1 ACC ACA AGG CAG ACA TTA GCT CCT and AAG GTA GTT CAG GGC ACT GTT CGT; SLAP1 TGC AGC TGC CAG ACA CAA AGA TTG and TCT GGA CCT TCC TGC AAA CTG GAT; GAPDH TGT GAG GTG ACC GCA TCT TCT TGT and ACG ACA TAC TCA GCA CCA GCA TCA. PCR was run at 95°C for 4 min followed by 25 cycles each at 95°C, 60°C, and 72°C for 30 sec at each temperature. The amplified products were separated on 1.3% agarose gel and stained with ethidium bromide for quantitation. Transcript levels of Dok1, DUSP1, SLAP1, and 18S rRNA (an internal control to calculate fold induction) were assayed by real-time PCR with the following primers and probes: DUSP1 ACCACAAGGCAGACATTAGCTCCT, AAACACCCTTCCTCCAGCATCCTT, probe FAM- TCAACGAGGCGATTGACTTTATAGACTCCA -TAMRA; SLAP1 ATCCAGTTTGCAGGAAGGTCCAGA, TAGGATGCGATGCTTTCCCGAAGA, probe FAM- AGAGAACCCACTCAGAGTGGACGAAT -TAMRA.
2.4. Cloning and mutation of a genomic SLAP and Dok1 promoters
RBL 2H3 cells were washed twice with 1 X phosphate buffered saline. Isolation and purification of chromosomal DNA was performed according to the manufacturer’s protocol. The desired promoter regions of chromosomal DNA were amplified by PCR with the following primers: Dok1 (region −2 to −2235, numbers indicate bp from transcription start site), AACGATATCTGCCCCATTCGCCGGCTCGC and ATGGATCCGCGATTCCTTCCGCGCTTCCT; SLAP1 (region −1 to −441), AACTCGAGAGGATTCCTGCATCCGCCTCT and AACAAGATCTACTGGAAAGGTAGAAAGCCTCC; the putative SLAP1 GRE region (+20721 to 21064), CGCTCGAGAACCCCAGCCAGAAAACAATGA and AAGCTAGCCCTCAGTACACACATACTGGAG. Mutagenesis of the putative SLAP1 GRE was performed by QuikChange Site-Directed Mutagenesis Kit with the following oligonucleotides: CTTCTGAAGAGGAATTAAGTAtcTCAgGTCCCAACCACTTCCTCCC and GGGAGGAAGTGGTTGGGACcTGAgaTACTTAATTCCTCTTCAGAAG (lower case letters indicate mutations). All amplified DNA sequences were confirmed by sequencing and cloned into the pGL4 luciferase reporter vector.
2.5. Dual luciferase reporter assay
RBL 2H3 cells (2 × 106 cells) were transfected with 2 μg pGL4.10 luciferase vector or with its SLAP1 or Dok1 constructs along with 0.5 μg pGL4.74TK Renillar luciferase as an internal control. Cells (2 × 105 cells) were incubated for 18 h in 24-well plates before addition of dexamethasone or other glucocorticoids individually or in combination with the GR antagonist RU486. Four hours later cells were washed with PBS and lysed in 1 X passive lysis buffer (Promega) for assay of firefly and Renillar luciferase activities in a Wallac Victor2 multiplate reader as prescribed by the manufacturer (Promega). Values were calculated as the ratio of firefly/Renillar luminescence (Fig. 3) or as fold increase over basal value obtained in the absence of glucocorticoid (Figs. 3 and 4).
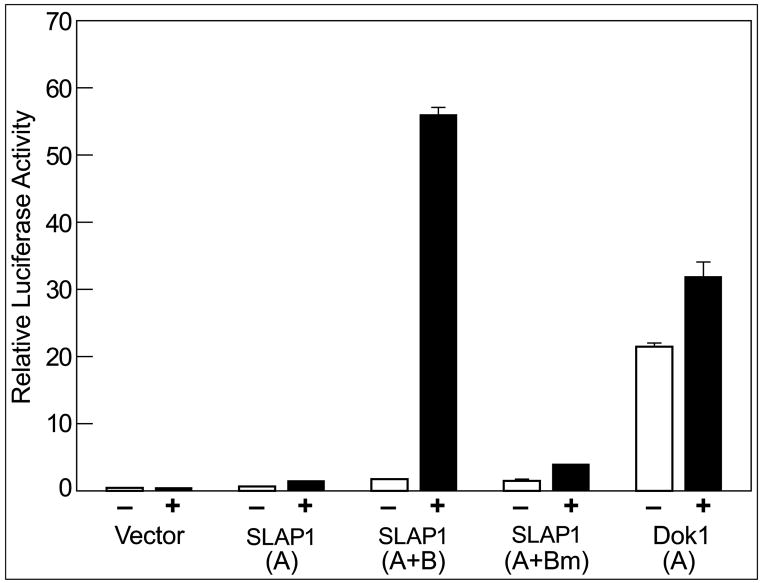
Fig. 3.
SLAP1 GRE luciferase reporter gene construct is activated by dexamethasone. RBL-2H3 cells were co-transfected with Renilla luciferase and the firefly luciferase vector with or without inclusion of the indicated gene elements. After 18 h, the cells were treated with vehicle or dexamethasone for 4 hours and harvested to measure luciferase activities. The ratios of the two luciferase activities (firefly/Renilla luciferase activity) are indicated. Key to gene elements (see Fig. 2): SLAP1 (A), region A of SLAP1; SLAP1 (A+B), region A plus B of SLAP1; SLAP1 (A+Bm), region A plus B with mutated GRE; Dok1 (A), region A of Dok1. See Materials and Methods sequence of mutated SLAP1 GRE.
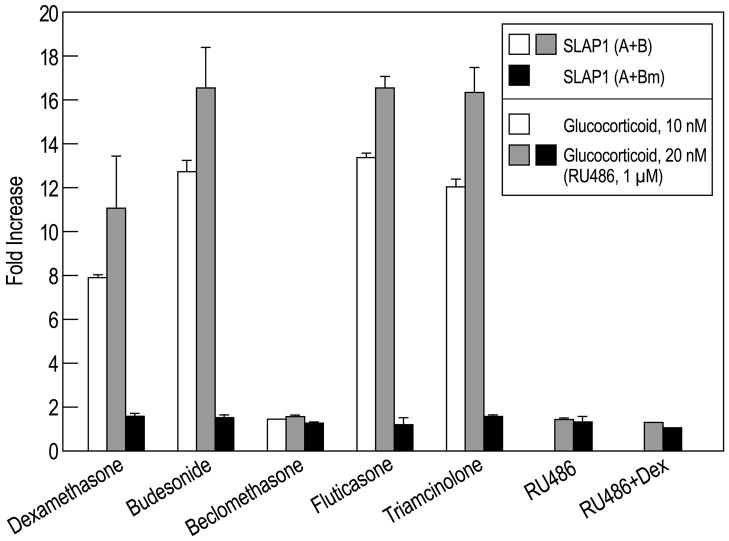
Fig. 4.
Additional evidence that SLAP1 GRE luciferase reporter gene is activated through GR. RBL-2H3 cells were co-transfected with Renilla luciferase and the firefly luciferase vector that contained either SLAP1 (A+B) or mutated SLAP1 (A+Bm). After 18 h, the indicated glucocorticoids (10 or 20 nM) were added with or without the GR antagonist RU486 (1 μM). Four hours later cells were lysed to measure luciferase activities. The ratio of the two luciferase activities (firefly/Renilla luciferase activity) in the absence of glucocorticoid was the denominator to calculate fold increase in activity in the presence of glucocorticoids.
2.6. Chromatin immunoprecipitation assay
RBL-2H3 cells (107 cells/20 ml/10 cm dish) were incubated with 100 nM dexamethasone or vehicle in growth medium for 30 minutes. Cells were then fixed with formaldehyde (1% final concentration) at 37°C for 5 minutes and washed twice with cold PBS before addition of 2 ml of lysis buffer (10 M Tris, pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail, and 0.1 mM fresh phenylmethanesulfonylfluoride). Residue was detached by scraping and, after centrifugation, resuspended in 1 ml lysis buffer. The suspension was sonicated (maximal setting) with a microtip ten times for 10 sec to ensure fragmentation of chromatin. The suspension was clarified by centrifugation (12000 × g, 15 min) before addition of 2 μg of antibody against GR or pre-serum and then kept at 4°C for 18 h. Immunoprecipitat ed GR/GRE complex was recovered by incubation with protein-A conjugated magnetic beads for 4 hours. The beads were washed twice with the lysis buffer, a lysis buffer containing 0.3 M NaCl, a lithium buffer (10 nM Tris, pH8.0, 0.2 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and a TET buffer (10 mM Tris, pH 8.0, 1 mM EDTA and 0.2% Triton X-100). The beads were then incubated at 65°C for 18 h with 100 μl of a reverse buffer (10 mM Tris, pH8.0, 1 mM EDTA, 0.3% SDS and 1 μg/μl protease K) to elute and separate GRE DNA and protein. DNA was extracted from this buffer by phenol/chloroform extraction, precipitated with ethanol, and dissolved in 50 μl water. SLAP1 GRE that was present in the extracted DNA and in DNA recovered from total cell lysate before immunoprecipitation were measured by real-time PCR with the primer pair, GTTCCTTCCTGAATGGCTCTCAGT and TCATACCATGTGGCAGACTCCCTT, and a probe, FAM- TGGGAGGAAGTGGTTGGGACATGAT -TAMRA. The ratio of immunoprecipitated and total GRE was calculated and data are presented as fold increase in this ratio in dexamethasone treated cells as compared to vehicle treated cells.
3. Results
3.1. Dexamethasone increases expression of Dok1, DUSP1, and SLAP1 transcripts by different mechanisms in RBL-2H3 cells
As noted earlier, the anti-inflammatory effects of glucocorticoids have been attributed in part to the upregulation of inhibitory molecules such as Dok1, DUSP1, and SLAP1 in mast cells by mechanisms that remain undefined. To examine these mechanisms we first reassessed the kinetics of induction of the inhibitory regulators by dexamethasone in RBL-2H3 cells. Dexamethasone increased transcript levels of all three in agreement with previous observations (Kassel et al., 2001; Hiragun et al., 2005; Hiragun et al., 2006). Levels of Dok1, DUSP1 and SLAP1 transcripts reached a maximum by 2 h after addition of dexamethasone (Fig. 1A to C). Dok1 and DUSP1 transcripts decreased thereafter whereas levels of SLAP1 transcripts remained elevated. The decline of DUSP1 was relatively rapid and approached basal levels by 6 h.
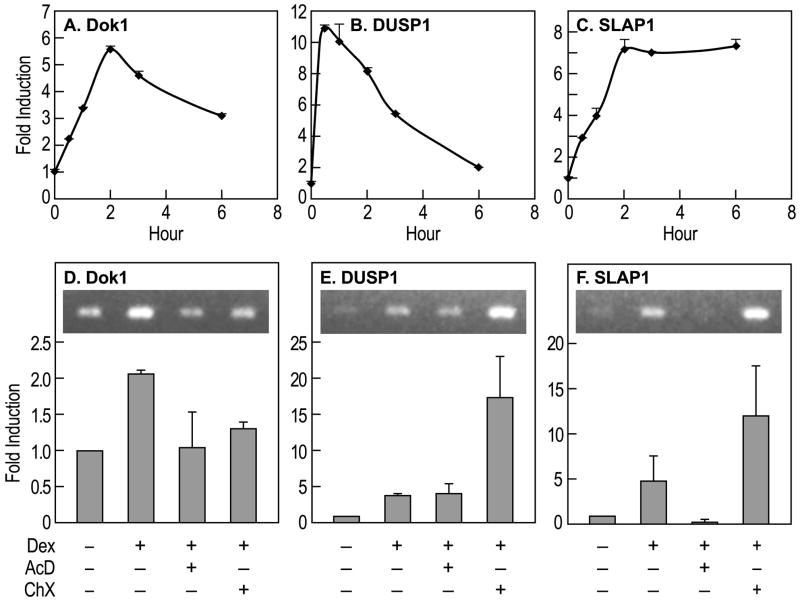
Fig. 1.
Upregulation of Dok1, DUSP1, and SLAP1 gene transcripts by dexamethasone and the effects of actinomycin D and cycloheximide. A–C, Dok1, DUSP1, and SLAP1 transcripts were measured by real time PCR at the indicated times after the addition of 100 nM dexamethasone (Dex) to RBL-2H3 cell cultures. D–F, Actinomycin D (AcD, 100 ng/ml) or cycloheximide (ChX, 1 μg/ml) were added 20 min before addition of dexamethasone and transcript levels were determined 4 h thereafter. Data are from two separate experiments (mean ± S.D.) and typical ethidium bromide-stained gels are shown in the insets.
We next investigated the effects of an RNA polymerase inhibitor, actinomycin D, and the protein synthesis inhibitor, cycloheximide, on the increase in transcript levels in dexamethasone-treated cells (Fig. 1D to F). The increase in Dok1 transcripts was inhibited by both actinomycin D and cycloheximide to indicate that both gene transcription and protein synthesis are involved in the upregulation of Dok1 (Fig. 1D). In contrast to Dok1, the increase in DUSP1 mRNA was not inhibited by actinomycin D but was enhanced by cycloheximide (Fig. 1E). A possible scenario is that suppression of dexamethasone-induced synthesis of mRNA-destabilizing tristetraproline (see Introduction) by cycloheximide might enhance stability of DUSP1 mRNA. DUSP1 mRNA contains two copies of AU-rich elements in the 3′ untranslated region and are targets for tristetraprolin (Lin et al., 2008; Emmons et al., 2008). In contrast to Dok1 and DUSP1, the increase in SLAP1 transcripts was blocked by actinomycin D and augmented by cycloheximide (Fig. 1F). Apparently, this increase is dependent on de novo synthesis of SLAP1 mRNA and on mechanism(s) that require protein synthesis. Unlike DUSP1 mRNA we found no AU-rich tristetraprolin recognition sites in SLAP1 mRNA.
3.2. Luciferase reporter assay of responsiveness of SLAP1 and Dok1 GREs to dexamethasone
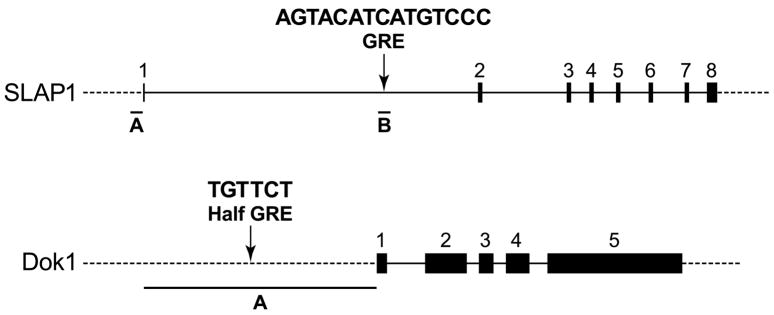
The inhibitory effects of actinomycin D on production of Dok1 and SLAP1 transcripts raise the possibility that these two genes are regulated, at least in part, through GREs. The MULAN program (http://mulan.dcode.org/) (Loots and Ovcharenko, 2007) was used to search for predicted GREs that are evolutionary conserved in human, mouse and rat in the Dok1, DUSP1, and SLAP1 genes. A predicted GRE site was present in the first intron of SLAP1 gene and a half-GRE was detected 1 kb upstream from transcription start site of the Dok1 gene (Fig. 2). However, no such sites were found within 10 kb upstream or downstream of the DUSP1 gene which was consistent with the lack of effect of actinomycin D on DUSP1 transcript levels (Fig.1D). The SLAP1 GRE is notable because of its location. This GRE and indeed the whole SLAP1 gene reside within the thyroglobulin gene which is transcribed in the opposite direction to the SLAP1 gene (Meijerink et al., 1998). Therefore, this GRE is a candidate regulator for both genes as thyroglobulin is also upregulated by dexamethasone in RBL-2H3 cells (Hiragun, T. and Beaven, M. A., unpublished data).
Fig. 2.
Organization of Dok1 and Slap1 genes to indicate location of GRE and half GRE. Upper panel: exons and introns are indicated by the numbered boxes and solid lines respectively. Dashed lines indicate non-transcribed regions. Regions that were amplified and cloned for this study are delineated and designated as A and B. These regions are highly conserved in human, mouse, and rat. Region A of the SLAP1 gene (441 bp) is a basal transcription element immediately upstream of exon 1 and region B (344 bp) contains GRE. Region A of Dok1 (2235 bp) embraces part of the promoter region that contains the half GRE. Lower panel: cloned sequences of regions A and B of the SLAP1 gene. Region B is in the reverse direction of chromosomal DNA and was inserted as such in the luciferase construct. Highly conserved binding sites for the indicated transcription factors and GR (GRE) are underlined. The + sign indicates that the consensus sequence for the binding site is in the same direction as that shown whereas the – sign indicates the reverse situation.
We amplified a region of 441 bp which was 1 bp upstream of exon 1 of the RBL-2H3 cell SLAP1 gene (region A of the SLAP1 gene, Fig. 2) and cloned it into pGL4.10 luciferase reporter vector. This region possessed some basal transcriptional activity because it elicited a modest increase in luciferase activity as compared to the vector control (Fig. 3). The upstream 344 bp region that contained the GRE (region B of SLAP1 gene, Fig. 2) was also amplified and inserted into the luciferase construct that contained region A. Dexamethasone treatment stimulated the promoter activity of this construct more than 30-fold. Mutation of the GRE (see Materials and methods) resulted in virtual loss of luciferase activity in response to dexamethasone (Fig. 3).
With respect to Dok1, a region that contained the one conserved half GRE upstream from the transcription start site (see Fig. 2) was amplified and inserted into the luciferase vector. This region excluded several other non-conserved half GREs. With this construct, basal activity before treatment with dexamethasone was relatively high. Dexamethasone treatment resulted in a modest 1.5 fold activation of lucifierase activity (Fig. 3). These results are consistent with the relatively high basal levels of Dok1 mRNA in the absence of dexamethasone (inset Fig. 1D) and the notion that GR interactions with half GREs operate only in conjunction with other regulatory sites (Schoneveld et al., 2004).
3.3. The SLAP1 luciferase reporter gene is activated through GR
The SLAP1 luciferase reporter gene that contained both the A and the GRE-containing B elements (Fig. 2) was activated by relatively low concentrations (10 nM and 20 nM) of budesonide, fluticasone, triamcinolone as well as dexamethasone (Fig. 4, open and grey bars). These concentrations were chosen because they were comparable to the reported values for EC50 (concentration required for 50% of maximum response) when these compounds were tested for their GRE transactivating activity in vitro (Jaffuel et al., 2000). Beclomethasone was inactive at these concentrations (Fig. 4) but it is reported to have relatively weak transactivation activity as compared to its transrepression activity. Moreover, beclomethasone had no demonstrable transactivation activity when assayed by upregulation of tyrosine aminotransferase activity in cells (Jaffuel et al., 2000).
The activation of the SLAP1 luciferase reporter gene by dexamethasone was blocked by the GR antagonist, RU486 (Fig. 4, right-hand columns). Furthermore, activation in response to all glucocorticoids was impaired by mutation of the GRE B element (Fig. 4, SLAP1 (A+Bm), solid bars). Collectively these data imply that activation of the luciferase reporter gene was dependent on interaction of ligand-activated GR with GRE.
3.4. Confirmation that ligand-bound GR interacts directly with SLAP1 GRE
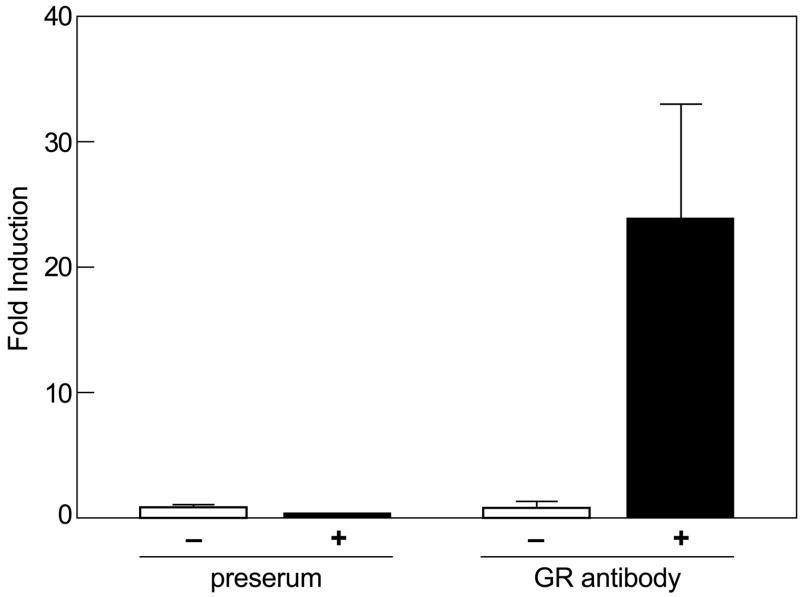
The above data indicated that the SLAP1 gene contained a fully functional GRE. To verify that this GRE is indeed the binding site for ligand-activated GR, chromatin immunoprecipitation was performed with anti-GR antibody. The data indicated a 26 fold-increase in the amount of precipitated GR/GRE complex in dexamethasone-treated cells as compared non-treated cells (Fig. 5). No reaction was observed with pre-immune serum.
Fig. 5.
ChIP analysis reveals direct interaction of GR with SLAP1 GRE. RBL-2H3 cells were incubated with 100 nM dexamethasone for 30 minutes, fixed with 1% formaldehyde, lysed, and processed for ChIP analysis with anti-GR antibody or pre-serum (see Materials and methods). The quantities of the SLAP1 GREs in the precipitated DNA fragments were measured by real time PCR. The data are presented as the increase in ratio of immunoprecipitated SLAP1 GRE and total SLAP1 GRE in whole cell lysate. Values are mean ± SD from two separate experiments.
4. Discussion
Here we demonstrate that the enhanced transcription of the SLAP1 gene in response to dexamethasone results, at least in part, to direct interaction of ligand-activated GR with the GRE in the first intron. The mechanisms for upregulation of Dok1 and DUSP1 were less clear although a half GRE may participate in regulating transcription of the Dok1 gene and increased message stability may account for the upregulation of DUSP1.
With respect to SLAP1, the GRE in the SLAP1 gene (Fig. 2, segment B) lies within a region that is conserved in human, mouse, and rat. This region contains not only the GRE but also nearby transcriptional binding sites for ARP1, NERF, ETS1, AFP1, and USF some of which, such as ETS1, are known to act co-operatively with the GR/GRE complex (Mullick et al., 2001). Further studies are needed to determine if this is true for the SLAP1 gene.
Except for lymphocytes and platelets relatively little is known about SLAP1 although it is expressed at the mRNA level in many tissues (Pandey et al., 1995; Meijerink et al., 1998). SLAP1 contains an amino-terminal myristoylation site that permits interaction of SLAP with cell membranes, nearby adjacent Src homology (SH)3 and SH2 domains, and a unique carboxy-terminal domain that can interact with c-Cbl. SLAP1 has been most intensively studied in lymphocytes. Studies with SLAP1-deficient mice have indicated that SLAP1 plays a crucial role in thymocyte and development and positive selection (Sosinowski et al., 2001). SLAP1 was initially characterized as a negative regulator of T cell receptor and B cell receptor signaling (Sosinowski et al., 2000; Tang et al., 1999; Dragone et al., 2006; Myers et al., 2006) and of platelet-derived growth factor-induced mitogenesis in platelets (Roche et al., 1998; Sirvent et al., 2008). In particular, SLAP1 acts cooperatively with c-Cbl to downregulate expression of T cell and B cell receptors (Myers et al., 2006; Dragone et al., 2006) and associates with several proteins that are essential for TCR-mediated signaling (Sosinowski et al., 2000) in a manner that is dependent on phosphorylation of the SLAP1 SH2 domain (Tang et al., 1999). These proteins include several relevant to FcεRI signal transduction such as Syk, LAT, and Vav. The more recently described SLAP family member, SLAP2 (Pandey et al., 2002; Holland et al., 2001), also interacts with Syk and its cognate ZAP70 and promotes their degradation in an SH2-dependent manner in Jurkat T cells (Loreto et al., 2002). As we have shown, SLAP1 suppresses FcεRI-mediated signaling events through Syk and LAT in mast cells (Hiragun et al., 2006) and future research may reveal other examples of negative regulation of tyrosine kinase-dependent receptor systems by SLAP proteins.
The upregulation of SLAP1 by dexamethasone in mast cells occurs at remarkably low concentrations of 5 to 10 nM. SLAP2, in contrast, is minimally expressed at the mRNA level in RBL-2H3 cells even after treatment with dexamethasone (our unpublished observations). The present studies clarify the mechanism for the upregulation of SLAP1 at a transcriptional level. Collectively, our studies support the notion that glucocorticoids suppress mast cell-mediated inflammatory reactions not only by suppressing cytokine gene transcription but also by increasing expression of inhibitory regulators of signaling which may have implications in the design of novel glucocorticoid receptor agonists directed towards minimizing gene activation and maximizing gene repression (Stahn et al., 2007).
Acknowledgments
The authors thank Drs Tae-Young Rho and Keji Zhao (NHLBI) for helpful advice on the ChIP assay. This work was supported by the Intramural research program of the National Heart, Lung, and Blood Institute.
ABBREVIATIONS
- Dok
downstream of tyrosine kinase
- DUSP
dual specificity protein phosphatase otherwise known as MAP kinase phosphatase
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- LAT
linker for activation of T cells
- MAP kinases
Mitogen activated protein kinases
- NTAL
non-T cell activation linker
- PL
phospholipase
- SLAP
Src-like adaptor protein
- TCR and BCR
T and B cell receptors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrade MV, Hiragun T, Beaven MA. Dexamethasone suppresses antigen-induced activation of phosphatidylinositol 3-kinase and downstream responses in mast cells. J Immunol. 2004;172:7254–7262. doi: 10.4049/jimmunol.172.12.7254. [DOI] [PubMed] [Google Scholar]
- Berenstein EH, Garcia-Gil M, Siraganian RP. Dexamethasone inhibits receptor-activated phosphoinositide breakdown in rat basophilic leukemia (RBL-2H3) cells. J Immunol. 1987;138:1914–1918. [PubMed] [Google Scholar]
- Clark AR. Anti-inflammatory functions of glucocorticoid-induced genes. Mol Cell Endocrinol. 2007;275:79–97. doi: 10.1016/j.mce.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Daeron M, Sterk AR, Hirata F, Ishizaka T. Biochemical analysis of glucocorticoid-induced inhibition of IgE-mediated histamine release from mouse mast cells. J Immunol. 1982;129:1212–1218. [PubMed] [Google Scholar]
- Dragone LL, Myers MD, White C, Gadwal S, Sosinowski T, Gu H, Weiss A. Src-like adaptor protein (SLAP) regulates B cell receptor levels in a c-Cbl-dependent manner. Proc Natl Acad Sci U S A. 2006;103:18202–18207. doi: 10.1073/pnas.0608965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons J, Townley-Tilson WH, Deleault KM, Skinner SJ, Gross RH, Whitfield ML, Brooks SA. Identification of TTP mRNA targets in human dendritic cells reveals TTP as a critical regulator of dendritic cell maturation. RNA. 2008;14:888–902. doi: 10.1261/rna.748408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nat Rev Immunol. 2006;6:218–230. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- Hiragun T, Peng Z, Beaven MA. Cutting Edge: Dexamethasone negatively regulates Syk in mast cells by up-regulating Src-like adaptor protein. J Immunol. 2006;177:2047–2050. doi: 10.4049/jimmunol.177.4.2047. [DOI] [PubMed] [Google Scholar]
- Hiragun T, Peng Z, Beaven MA. Dexamethasone up-regulates the inhibitory adaptor protein Dok-1 and suppresses downstream activation of the mitogen-activated protein kinase pathway in antigen-stimulated RBL-2H3 mast cells. Mol Pharmacol. 2005;67:598–603. doi: 10.1124/mol.104.008607. [DOI] [PubMed] [Google Scholar]
- Holland SJ, Liao XC, Mendenhall MK, Zhou X, Pardo J, Chu P, Spencer C, Fu A, Sheng N, Yu P, Pali E, Nagin A, Shen M, Yu S, Chan E, Wu X, Li C, Woisetschlager M, Aversa G, Kolbinger F, Bennett MK, Molineaux S, Luo Y, Payan DG, Mancebo HS, Wu J. Functional cloning of Src-like adapter protein-2 (SLAP-2), a novel inhibitor of antigen receptor signaling. J Exp Med. 2001;194:1263–1276. doi: 10.1084/jem.194.9.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol Reprod. 2005;72:1290–1296. doi: 10.1095/biolreprod.105.040014. [DOI] [PubMed] [Google Scholar]
- Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006;117:522–543. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Jaffuel D, Demoly P, Gougat C, Balaguer P, Mautino G, Godard P, Bousquet J, Mathieu M. Transcriptional potencies of inhaled glucocorticoids. Am J Respir Crit Care Med. 2000;162:57–63. doi: 10.1164/ajrccm.162.1.9901006. [DOI] [PubMed] [Google Scholar]
- Jeong HJ, Na HJ, Hong SH, Kim HM. Inhibition of the stem cell factor-induced migration of mast cells by dexamethasone. Endocrinology. 2003;144:4080–4086. doi: 10.1210/en.2003-0115. [DOI] [PubMed] [Google Scholar]
- Kassel O, Sancono A, Kratzschmar J, Kreft B, Stassen M, Cato AC. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 2001;20:7108–7116. doi: 10.1093/emboj/20.24.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Hammer M, Mages J. DUSP meet immunology: dual specificity MAPK phosphatases in control of the inflammatory response. J Immunol. 2006;177:7497–7504. doi: 10.4049/jimmunol.177.11.7497. [DOI] [PubMed] [Google Scholar]
- Lin NY, Lin CT, Chang CJ. Modulation of immediate early gene expression by tristetraprolin in the differentiation of 3T3-L1 cells. Biochem Biophys Res Commun. 2008;365:69–74. doi: 10.1016/j.bbrc.2007.10.119. [DOI] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. Mulan: multiple-sequence alignment to predict functional elements in genomic sequences. Methods Mol Biol. 2007;395:237–254. [PMC free article] [PubMed] [Google Scholar]
- Loreto MP, Berry DM, McGlade CJ. Functional cooperation between c-Cbl and Src-like adaptor protein 2 in the negative regulation of T-cell receptor signaling. Mol Cell Biol. 2002;22:4241–4255. doi: 10.1128/MCB.22.12.4241-4255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijerink PH, Yanakiev P, Zorn I, Grierson AJ, Bikker H, Dye D, Kalaydjieva L, Baas F. The gene for the human Src-like adaptor protein (hSLAP) is located within the 64-kb intron of the thyroglobulin gene. Eur J Biochem. 1998;254:297–303. doi: 10.1046/j.1432-1327.1998.2540297.x. [DOI] [PubMed] [Google Scholar]
- Mullick J, Anandatheerthavarada HK, Amuthan G, Bhagwat SV, Biswas G, Camasamudram V, Bhat NK, Reddy SE, Rao V, Avadhani NG. Physical interaction and functional synergy between glucocorticoid receptor and Ets2 proteins for transcription activation of the rat cytochrome P-450c27 promoter. J Biol Chem. 2001;276:18007–18017. doi: 10.1074/jbc.M100671200. [DOI] [PubMed] [Google Scholar]
- Myers MD, Sosinowski T, Dragone LL, White C, Band H, Gu H, Weiss A. Src-like adaptor protein regulates TCR expression on thymocytes by linking the ubiquitin ligase c-Cbl to the TCR complex. Nat Immunol. 2006;7:57–66. doi: 10.1038/ni1291. [DOI] [PubMed] [Google Scholar]
- Nakamura R, Okunuki H, Ishida S, Saito Y, Teshima R, Sawada J. Gene expression profiling of dexamethasone-treated RBL-2H3 cells: induction of anti-inflammatory molecules. Immunol Lett. 2005;98:272–279. doi: 10.1016/j.imlet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Pandey A, Duan H, Dixit VM. Characterization of a novel Src-like adapter protein that associates with the Eck receptor tyrosine kinase. J Biol Chem. 1995;270:19201–19204. doi: 10.1074/jbc.270.33.19201. [DOI] [PubMed] [Google Scholar]
- Pandey A, Ibarrola N, Kratchmarova I, Fernandez MM, Constantinescu SN, Ohara O, Sawasdikosol S, Lodish HF, Mann M. A novel Src homology 2 domain-containing molecule, Src-like adapter protein-2 (SLAP-2), which negatively regulates T cell receptor signaling. J Biol Chem. 2002;277:19131–19138. doi: 10.1074/jbc.M110318200. [DOI] [PubMed] [Google Scholar]
- Rivera J, Gilfillan AM. Molecular regulation of mast cell activation. J Allergy Clin Immunol. 2006;117:1214–1225. doi: 10.1016/j.jaci.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Roche S, Alonso G, Kazlauskas A, Dixit VM, Courtneidge SA, Pandey A. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr Biol. 1998;8:975–978. doi: 10.1016/s0960-9822(98)70400-2. [DOI] [PubMed] [Google Scholar]
- Sayed BA, Christy A, Quirion MR, Brown MA. The master switch: the role of mast cells in autoimmunity and tolerance. Annu Rev Immunol. 2008;26:705–739. doi: 10.1146/annurev.immunol.26.021607.090320. [DOI] [PubMed] [Google Scholar]
- Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Sirvent A, Leroy C, Boureux A, Simon V, Roche S. The Src-like adaptor protein regulates PDGF-induced actin dorsal ruffles in a c-Cbl-dependent manner. Oncogene. 2008;27:3494–3500. doi: 10.1038/sj.onc.1211011. [DOI] [PubMed] [Google Scholar]
- Smoak K, Cidlowski JA. Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Mol Cell Biol. 2006;26:9126–9135. doi: 10.1128/MCB.00679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci U S A. 2008;105:5745–5749. doi: 10.1073/pnas.0801551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinowski T, Killeen N, Weiss A. The Src-like adaptor protein downregulates the T cell receptor on CD4+CD8+ thymocytes and regulates positive selection. Immunity. 2001;15:457–466. doi: 10.1016/s1074-7613(01)00195-9. [DOI] [PubMed] [Google Scholar]
- Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like adaptor protein (SLAP) is a negative regulator of T cell receptor signaling. J Exp Med. 2000;191:463–474. doi: 10.1084/jem.191.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahn C, Lowenberg M, Hommes DW, Buttgereit F. Molecular mechanisms of glucocorticoid action and selective glucocorticoid receptor agonists. Mol Cell Endocrinol. 2007;275:71–78. doi: 10.1016/j.mce.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Stellato C. Post-transcriptional and nongenomic effects of glucocorticoids. Proc Am Thorac Soc. 2004;1:255–263. doi: 10.1513/pats.200402-015MS. [DOI] [PubMed] [Google Scholar]
- Tang J, Sawasdikosol S, Chang JH, Burakoff SJ. SLAP, a dimeric adapter protein, plays a functional role in T cell receptor signaling. Proc Natl Acad Sci U S A. 1999;96:9775–9780. doi: 10.1073/pnas.96.17.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]