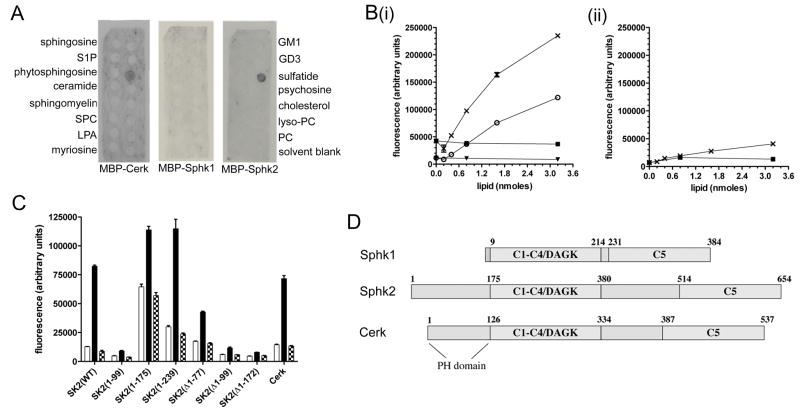
Figure 1. Sphk2 and Cerk bind sulfatide.
(A) Interaction of MBP-Cerk, MBP-Sphk1, and MBP-Sphk2 with lipids immobilised on Sphingostrip membranes. Identity of the lipid in each 100 nmole spot is indicated alongside the blots: S1P, sphingosine 1-phosphate; SPC, sphingosylphosphorylcholine; LPA, lysophosphatidic acid; GM1, monosialoganglioside type GM1; GD3, disialoganglioside type GD3; PC, phosphatidylcholine. No signal was observed with MBP control protein. (B) Binding of MBP-Sphk2 (i) or MBP-Sphk1 (ii), at 0.25 (○ and ▼) or 1 mg/mL (X and ■), to immobilised sulfatide (○ and X) or cerebroside (▼ and ■). (C) Binding of MBP fusion proteins (0.5 μg/mL) to wells coated with BSA only (open bars), 2 nmole sulfatide (closed bars), or 2 nmole cerebroside (hatched bars). Sphk2(1-99), Sphk2(1-175), and Sphk2(1-239) refer to the first 99, 175, and 239 residues of Sphk2, respectively, whilst Δ1-77, Δ1-99, and Δ1-172 indicate deletion of those residues. Results shown are mean and standard error of triplicate wells. (D) Domain structures of human Sphk1, Sphk2 and Cerk; numbers indicate primary amino acid sequence. C1–C5: sphingolipid kinase conserved domains; DAGK: diacylglycerol kinase active site.