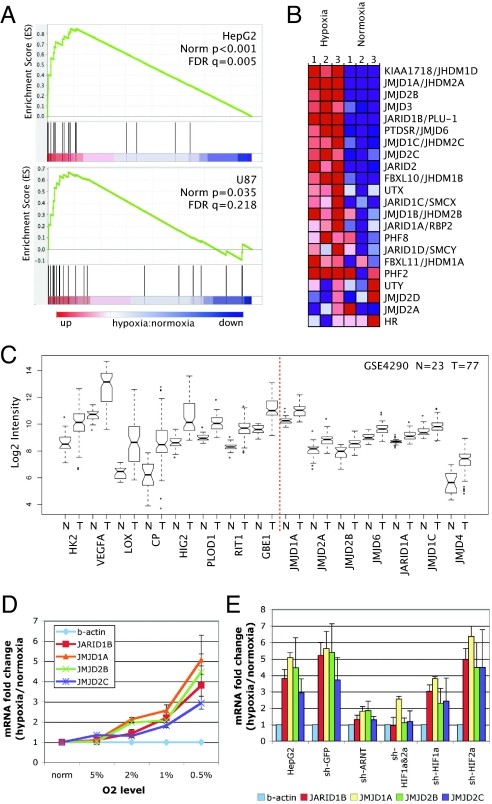
Fig. 4.
Hypoxia- and HIF-dependent up-regulation of JHDMs. (A) GSEA analysis of mRNA expression profiles for hypoxic (12 h) vs. normoxic cells with gene set containing all known JmjC histone demethylases in HepG2 and U87 cells. (B) Gene expression changes under hypoxia in HepG2 cells. (C) Expression dataset for Grade IV glioblastoma was interrogated for changes in mRNA levels of the JmjC family. The most highly up-regulated JmjC family member is shown for normal tissue (N) vs. tumor (T) samples. Known HIF-1 targets are on the left. Data represented as the median (bar), 25–75th percentiles (box), and range (whiskers). (D) Quantitative RT-PCR verification of up-regulated expression of HIF-1-bound JmjC genes under different levels of hypoxia. Results are expressed as mean ± SD after normalization to β-actin. (E) shRNA depletion of HIF-1α, HIF-2α, HIF1α and 2α, or ARNT was used to determine HIF-dependent up-regulation of JmjC proteins. Data represent fold change comparing hypoxic with normoxic samples (mean ± SD).