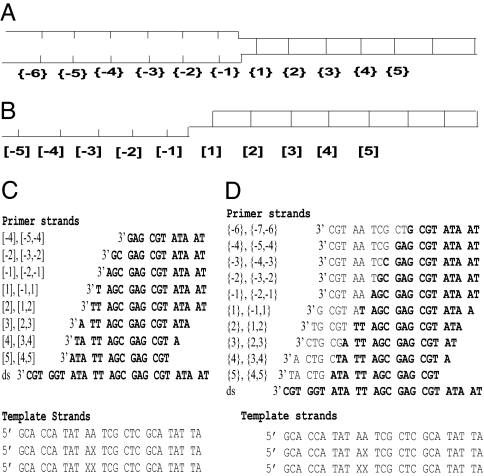
Fig. 1.
Nomenclature and sequences of the DNA constructs used. (A and B) The overall design of the forked (A) and the P/T (B) constructs. Negative numbers designate residue positions in ssDNA regions, and positive numbers correspond to dsDNA base pair positions. {} designate positions within the forked constructs; [] designate positions within the P/T constructs. (C and D) The sequence of the template (Lower) and corresponding primer (Upper) strands for P/T (C) and forked (D) constructs. X represents the position of 2-AP residue. Complementary base pairs in the various top strands are in bold. Each construct is designated by construct type (P/T or forked) and the position of the probe residues with respect to the single-stranded/double-stranded junction; the nomenclature used to designate the various duplex constructs is shown at the left of each nontemplate sequence.