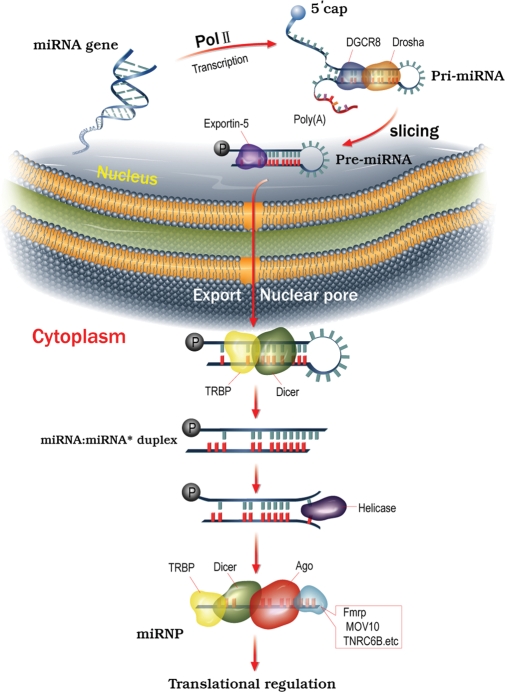
Figure 1.
miRNA biogenesis. Genes encoding miRNAs are initially transcribed by RNA polymerase II or III to generate the pri-miRNA transcripts within the nucleus. The stem-loop structure of the pri-miRNA is recognized and cleaved on both strands by a microprocessor complex, which consists of the nuclear RNase III enzyme Drosha and an RNA-binding protein, DGCR8, to yield a pre-miRNA 60–70 nt in length. The pre-miRNA is then exported from the nucleus through a nuclear pore by exportin-5 in a Ran-GTP-dependent manner and processed in the cytoplasm by the RNase III Dicer–TRBP. Sliced RNA strands are further unwound by an RNA helicase. One strand of the miRNA/miRNA* or siRNA duplex (the antisense, or guide strand) is then preferentially incorporated into the RISC (or miRNP for miRNAs) and will guide the miRNP to a target mRNA in a sequence-specific manner. Once directed to a target mRNA, the RISC can mediate translational regulation by inhibiting the initiation or elongation step or through destabilization of the target mRNA. Alternatively, miRNAs may also upregulate translation of target mRNAs under certain conditions.