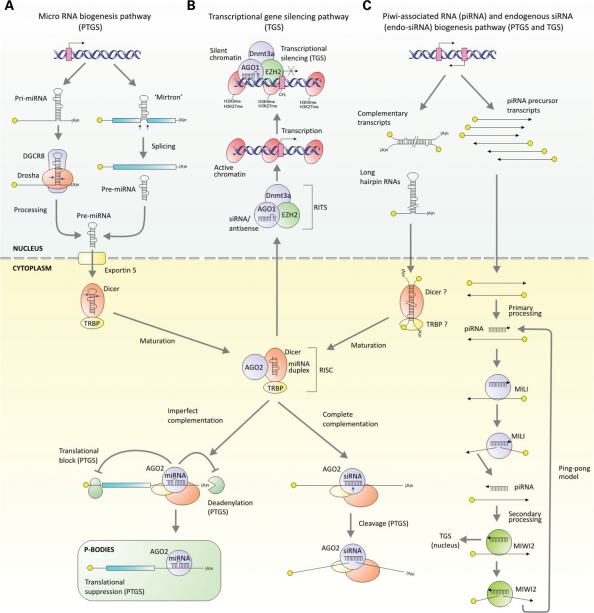
Figure 1.
Mammalian RNAi regulatory pathways. (A) miRNAs are encoded in pri-miRNAs (124,125). These ∼100 nt inverted repeat motifs are usually found embedded once or multiple times within coding or non-coding RNA Pol II-derived transcripts (126). Pri-miRNAs are first processed in the nucleus where their hairpin-like structures are recognized and cleaved by the RNase III enzyme Drosha together with DiGeorge critical region 8 protein (DGCR8), to produce shorter hairpin duplexes known as a 70–80 nt pre-miRNAs (127,128). For a small minority of miRNAs, short intronic sequences, referred to ‘mirtrons’, can be directly processed by the spliceosome into pre-miRNA-like hairpins without requiring Drosha cleavage (30,129). Spliced lariats are de-branched and likely produce functional pre-miRNAs for export. Pre-miRNAs are exported from the nucleus to the cytoplasm by the exportin-5 (130,131) followed by recognition and cleavage by a second RNase III enzyme, Dicer and its partner, TAR RNA-binding protein (TRBP), to produce a ∼22 bp, staggered miRNA/miRNA* duplex with 2 nt 3′ overhangs (132–134). Dicer/TRBP, loads one of the strands, the ‘guide strand’, into a RISC consisting in its simplest form of Ago2 (135,136) directing cleavage of translational suppression of cognate RNA. (B) The mechanism of TGS is poorly understood in mammals but is thought to include a complex consisting (RITS) of Ago1, (and possibly Ago2) a polycomb group component, enhancer of zeste 2 (EZH2) and DNA methyltransferase 3a (Dnmt3a) (21,23,137). Moreover, TGS may require the presence of low-copy promoter-derived transcripts to direct silent heterochromatin marks (H3K9 and/or H3K27 methylation) and DNA methylation at the targeted locus (138). (C) Endogenous siRNAs are derived from long hairpin sequences and complementary transcripts which are processed by Dicer into siRNAs. piRNAs are 24–31 nt short RNAs processed from single-stranded precursors derived from transposons or genomic repeat elements in the germline (13). In the ‘ping-pong model’, primary piRNAs interact with the Piwi protein MILI to cleave a transcript that generates a piRNA for incorporation into MIWI2, which in turn cycles back to produce new MILI-interacting piRNAs (24,25).