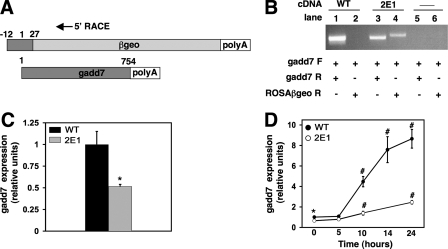
FIGURE 2.
Disruption of one gadd7 allele in 2E1 cells leads to decreased gadd7 RNA expression. A, schematic representation of gadd7-retroviral fusion transcript with 5′ RACE primer location and endogenous gadd7 transcript. B, directed PCR was performed on cDNA from WT (lanes 1 and 2) and 2E1 (lanes 3 and 4) cells to detect gadd7 expression (lanes 1 and 3) and fusion transcript (lanes 2 and 4). Negative control reactions (lanes 5 and 6) contained no cDNA. Forward (F) and reverse (R) primers for gadd7 were used in PCR for lanes 1, 3, and 5. Forward gadd7 primer and reverse primer for the proviral sequence (ROSAβgeo) were used in PCR for lanes 2, 4, and 6. C, basal gadd7 RNA expression in WT and 2E1 mutant cells was determined by qPCR and normalized to β-actin RNA expression. *, p < 0.05 for 2E1 versus WT. Data are expressed as mean ± S.E. for three independent experiments. D, WT and 2E1 mutant cells were treated with 500 μm palmitate for the indicated times and gadd7 expression was determined by qPCR and normalized to β-actin RNA expression. *, p < 0.05 for 2E1 versus WT; #, p < 0.01 for treated versus untreated WT cells and for treated 2E1 versus treated WT. Data are expressed as mean ± S.E. for three independent experiments.