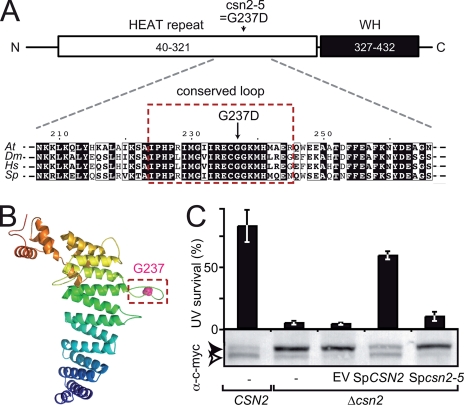
FIGURE 2.
The csn2-5 mutation modifies an evolutionarily conserved loop of CSN2. A, schematic representation of the CSN2 structural domains: N-terminal (N) HEAT repeat (residues 40–321) and C-terminal (C) winged helix (WH) domain (residues 327–432). The position and nature of the amino acid substitution found in csn2-5 mutant protein is shown. Below, an alignment of CSN2 proteins from Arabidopsis (Q8W207; residues 207–266), Drosophila (Q94899; residues 212–271), human (P61201; residues 211–270) and fission yeast (Q9HFR0; residues 213–272) around Arabidopsis Gly237 is shown. Residues boxed in red are highly conserved and belong to a predicted hairpin-like loop structure (framed in B). B, structural model of Arabidopsis CSN2 (residues 40–439) represented as ribbons and colored from the N to the C terminus in blue to red, respectively. Gly237 is highlighted as a pink sphere. C, the csn2-5 mutation abolishes CSN function in S. pombe as assessed by survival after UV-C exposure (50 kJ·m-2·s-1) relative to control. Wild-type CSN2 and mutant Δcsn2 strains expressing wild-type or mutant SpCSN2 were analyzed for complementation of the Δcsn2 UV hypersensitivity. Errors bars represent S.D. between three biological replicates. One representative experiment of two is shown. The level of RUB modification of c-Myc-tagged CUL1 (Pcu1p) expressed in the yeast strains was analyzed by immunoblot of total protein extracts using anti-c-Myc antibody. The open and filled arrowheads correspond to unmodified and RUB-modified cullins, respectively. EV, empty vector.