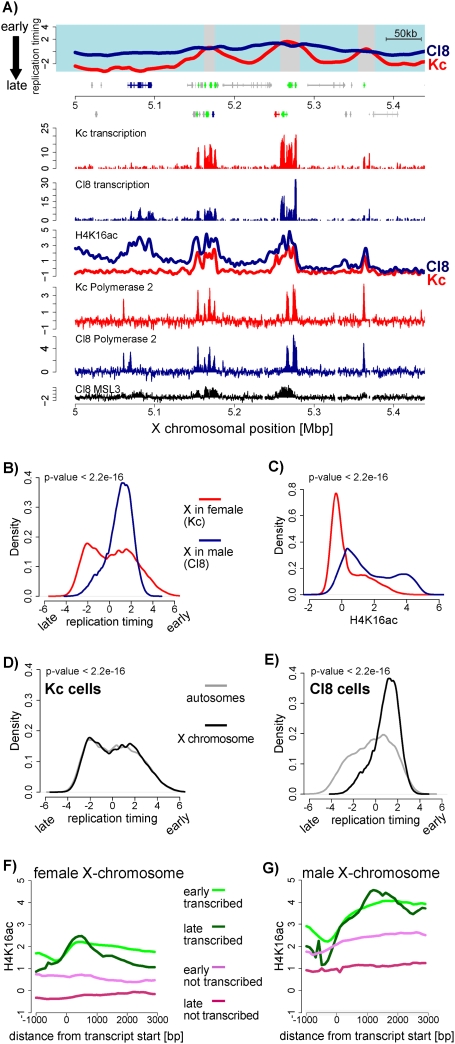
Figure 4.
Early replication timing of the male X chromosome. (A) Replication timing and transcription of a representative region on the X. Cl8 is shown in blue and Kc is shown in red (similar to Fig. 3A). In addition, H4K16ac and RNA polymerase II profiles are displayed for Kc (red line) and Cl8 (blue line) cells (Affymetrix tiling arrays). Previously published MSL3 binding data in Cl8 cells (Alekseyenko et al. 2006) are shown in black. (B) Quantitative comparison of replication timing between male and female X chromosomes. Shown is the frequency of array probes for any given replication timing value (X-axis) separately for the X in male (Cl8, blue line) and for the X in female (Kc, red line). This density plot shows that most of the male X replicates early (above zero), while the female X replicates similarly to autosomes. (C) Comparison of H4K16 acetylation for all X-linked array probes similar to B. This density plot shows that the male X is strongly enriched for H4K16 acetylation. (D,E) Density plot of replication timing on the X chromosome versus autosomes in female Kc cells (D) and in male Cl8 cells (E). P-values were calculated using the Wilcoxon rank-sum test. (F) Average signal for H4K16ac in Kc cells at genes. Genes were grouped according to replication timing and transcriptional activity into “active, early replicating,” (green line), “active, late replicating,” (dark-green line), “inactive, early replicating,” (violet line), and “inactive, late replicating” (red line). Genes were aligned at their start sites, and signals were averaged (see the Supplemental Material). (G) Similar analysis as in F for male cells (Cl8). Note that the male X chromosome contains higher H4K16 acetylation signals throughout active genes, but also in the gene body of early replicating nontranscribed genes.