Abstract
We recently described the assembly of fluorescent deoxyriboside monomers (“fluorosides”) into DNA-like phosphodiester oligomers (oligodeoxyfluorosides, or ODFs) in which hydrocarbon and heterocyclic aromatic fluorophores interact both physically and electronically. Here we report the identification of a multicolor set of water-soluble ODF dyes that display emission colors across the visible spectrum, and all of which can be simultaneously excited by long-wavelength UV light at 340−380 nm. Multispectral dye candidates were chosen from a library of 4096 tetramer ODFs constructed on PEG-polystyrene beads using a simple long-pass filter to observe all visible colors at the same time. We re-synthesized and characterized a set of 23 ODFs containing one to four individual chromophores, and included 2−3 spacer monomers to increase aqueous solubility and minimize aggregation. Emission maxima of this set range from 376 nm to 633 nm, yielding apparent colors from violet to red, all of which can be visualized directly. The spectra of virtually all ODFs in this set varied considerably from the simple combination of monomer components, revealing extensive electronic interactions between the presumably stacked monomers. In addition, comparisons of anagrams in the set (isomers having the same components in a different sequence) reveal the importance of nearest-neighbor interactions in the emissive behavior. Preliminary experiments with human tumor (HeLa) cells, observing two ODFs by laser confocal microscopy, showed that they can penetrate the outer cellular membrane, yielding cytoplasmic localization. In addition, a set of four distinctly-colored ODFs was incubated with live zebrafish embryos, showing tissue penetration, apparent biostability, and no apparent toxicity. The results suggest that ODF dyes may be broadly useful as labels in biological systems, allowing the simultaneous tracking of multiple species by color, and allowing visualization in moving systems where classical fluorophores fail.
Keywords: Fluorophore, reporter, exciplex, excimer, Stokes shift, DNA, library
Introduction
Fluorescence methods are among the most powerful classes of analytical research tools in the biosciences, and they are also among the most rapidly growing.1-8 Examples of new fluorescence methods and technologies that are under widespread use include laser confocal microscopy, single molecule microscopy, expression microarrays, real-time PCR assays (e.g., employing Taqman and molecular beacon probes), fluorescence in situ hybridization (FISH), chromosome painting, fluorescence activated cell sorting, two-photon dyes, near-IR dyes for tissue imaging, fluorescent chemosensors, fluorescence-based SNP scoring, green fluorescent protein and variants, and semiconductor quantum dots, among others. A major fraction of laboratories in the chemical, biological and biomedical sciences make use of fluorescence methods, and of fluorophores as reporters.
Despite the large amount of research directed at making new classes of fluorescent labels, commonly available organic fluorophores still suffer from a number of limitations. One of the most important limitations is that they have widely varied absorption maxima. If one wishes to perform fluorescence experiments using two or more dyes emitting at different wavelengths, several complications commonly arise. Laser-based instruments are usually quite limited in what dyes can be used, because the excitation is extremely narrow. If broadband illumination sources (e.g., mercury or xenon) are used, one must then use filters to avoid interference. For example, on an epifluorescence microscope one cannot easily view both fluorescein (green) and tetramethylrhodamine (TAMRA, orange-red) simultaneously because the light needed to excite TAMRA will interfere with fluorescein emission. Therefore, the two dyes are observed in separate experiments, using separate, costly excitation and emission filter sets that are specialized for each dye. For similar reasons, it is common that simple fluorescence instruments (such as microplate readers) are optimized for one dye only, or perhaps two if multiple light sources and/or filter sets are incorporated.
The limited photophysical properties of common dyes have an even more severe effect on the study of dynamic systems. In a rapidly moving biological sample, one does not have sufficient time to switch excitation and emission filters to image a second or third color. Thus observing multiple species move relative to one another, such as with membrane diffusion,9 for example, is typically limited to one color unless the motion is slow.
Some research groups have begun to address these limitations. Significant approaches include the development of quantum dots and FRET dye pairs. Semiconductor quantum dots (QDs)10-12 and other inorganic nanoparticles13 show multispectral emission (depending on size) but can be excited by a single short wavelength. However, preparation of well-defined, single conjugates can be more difficult with such large inorganic particles than for classical organic dyes. In addition, because of the passivating layers added to the surface of QDs, energy transfer and quenching can be inefficient as compared with small organic dyes.14,15 Moreover, their cell permeability can be limiting.12 Thus although QDs are commercially available, their use remains much less widespread than classical organic fluorophores. A second approach that addresses the spectral limitation is FRET dye pairs, which contain two fluorophores, a donor and an acceptor, wherein the same donor but different acceptors are used for Förster energy transfer.16 Such pairs have been useful in DNA sequencing, allowing sequencers to use a single laser excitation, with detection of four emission colors. With few exceptions,17 only the FRET mechanism has been generally used for such fluorophore dimers, and further complexity of electronic interactions has not been explored.
We have undertaken a program in which we assemble ∼3−5 fluorophores in a DNA-like chain, called an “oligodeoxyfluoroside” (ODF).18,19 Each fluorophore replaces the DNA base on a deoxyribose moiety,20,21 and a DNA synthesizer is used to assemble these oligomers, which have a negatively charged, water-soluble phosphodiester backbone like DNA. The fluorophores included in our libraries are typically simple aromatic hydrocarbons and heterocycles that can undergo multiple forms of energy transfer, including FRET, exciplex, excimer, H-dimer and other mechanisms.19 Our hypothesis is that the DNA backbone encourages close interaction of such flat aromatic species (as it was evolved to do for DNA bases), allowing for highly efficient electronic communication of excitation energy.22-25
In principle, such compounds might offer a solution to this photophysical limitation of classical fluorophores. If an ODF were to contain a chromophore that can be excited at one desired wavelength, it might then transduce its energy (by one of several possible energy transfer mechanisms) to longer wavelengths across the visible spectrum.18 Here we describe the evaluation of a large set of ODF tetramers for their ability to yield multispectral emission with a single excitation source and a single filter set. We report on a selected set of 23 differently-colored ODF reporters, all of which are excited by long-wavelength UV light, and we describe initial tests of their use in biological systems.
Experimental Section
Monomer syntheses
Syntheses of the four monomer deoxyribosides Y, E, B, and D (Fig. 1) were carried out as previously reported.18,20 They were converted to the 5’-dimethoxytrityl-3’-phosphoramidite derivatives as described. The spacer phosphoramidite (S) was purchased from Glen Research. Three of the fluorophore deoxyribosides (J, K, R) were not described previously; they were prepared from deoxyribose and the hydroxyethyl derivatives of the parent dyes24-26 as described below.
Figure 1.
Fluorescent deoxyriboside monomers employed as components of oligodeoxyfluoroside (ODF) dyes. Their one-letter abbreviations are shown. A nonfluorescent spacer monomer (S) is included.
General procedure for acetal formation from deoxyribose and hydroxyethyl dyes
An oven-dried two neck flask under argon purge was charged with the hydroxyethyl dye (1.0 equiv), silver oxide (1.5 equiv), Drierite (ca. 3 g), and Hoffer's chlorosugar (1.0 equiv). After sealing, sufficient anhydrous chloroform was introduced (5 to 10 mL) to allow the slurry to stir. The reaction vessel was wrapped in aluminum foil and allowed to stir 12 h, at which point an additional 0.5 equiv of chlorosugar was added. Total reaction time was 24 hours. The reaction mixture was filtered over a cotton plug and rinsed repeatedly with dichloromethane. Following evaporation of the solvent and column chromatography, a mixture of both alpha and beta anomers of deoxyribosides was isolated for J,K,R respectively. Separation was achieved following formation of the 5’-dimethoxytrityl derivatives.
General procedure for formation of free nucleosides J,K,R
To an oven dried round bottom flask under argon purge was added 2.5 mL sodium methoxide (25% by weight in methanol). The toluoyl protected nucleoside was dissolved in dry dichloromethane (10 mL) and introduced by syringe into the sodium methoxide solution. Deprotection was monitored by TLC. The reaction mixture was diluted with dichloromethane and rinsed with water. The organic layer was dried with sodium sulfate, evaporated under vacuum and purified by silica column chromatography (dichloromethane to 95% dichloromethane/5% methanol).
General procedure for formation of 5’-dimethoxytrityl derivatives of J,K,R
The free nucleoside was co-evaporated with dry pyridine (2 × 10 mL). To the reaction vessel was added dimethoxytrityl chloride (1.5 equiv), and N, N-diisopropylethylamine (5 equiv) and dry pyridine (10 mL). Progress of the reaction was monitored by thin layer chromatography. Excess dimethoxytrityl chloride was quenched with methanol and after 10 min, the reaction mixture was diluted with ethyl acetate. The organic layer was rinsed repeatedly with water, dried with sodium sulfate and reduced under vacuum. Column chromatography with a solvent gradient of hexanes : ethylacetate 3:1 to hexanes : ethylacetate 1:1 (with 5% triethylamine) provided the α-anomer as verified by ROESY and COSY experiments.
General procedure for formation of 3’-phosphoramidite derivatives of J,K,R
The 5’ dimethoxytrityl protected nucleoside was co-evaporated with toluene (3 × 10 mL). Diisopropylamine (1.05 eq), tetrazole (1.05 eq, solution in acetonitrile), bis(diisopropylamino)-2-cyanoethoxyphosphine (1.1 eq) and dichloromethane were added such that the final concentration of protected nucleoside was 0.1 M. The reaction mixture was stirred under argon for 2 hours removed from the vessel via syringe and filtered while injecting into an argon purged phosphoramidite bottle (Glen Research) for use on the DNA synthesizer.
Spectral data for nucleoside derivatives of J,K,R
5’,3’-O-bis-p-toluoyl-2’-deoxyribose-1’-α-acetal J
1H-NMR data (400.115 Hz), δ: 7.93 (m, 4H), 7.35 (d, 2H, J = 9.1 Hz), 7.29 (d, 1H, J = 15.8 Hz), 7.23 (m, 6H), 6.65 (d, 2H, J = 8.8 Hz), 6.54 (d, 2H, J = 27.7 Hz), 6.41 (d, 1H, J = 15.9 Hz), 5.55 (p, 1H, J = 3.5 Hz ), 5.28 (m, 1H), 4.52 (m, 2H), 4.38, dd, 1H, J = 8.2 Hz, J = 12.8 Hz), 3.90 (m, 1H), 3.59 (m, 3H), 2.99 (s, 3H), 2.55 (m, 1h), 2.39 (s, 3H), 2.37 (s, 3H), 2.36 (m, 2H). MS: calc'd 650.3 found 673.3 (ESI+, w/ Na+).
5’,3’-O-bis-p-toluoyl-2’-deoxyribose-1’-α-acetal K
1H-NMR data (400.115 Hz), δ: 7.84 (m, 4H), 7.26 (m, 3H), 7.16 (m, 4H), 6.60 (m, 2H), 6.49 (s, 1H), 6.41 (s, 1H), 6.31 (m, 1H), 5.49 (m, 1H), 5.32 (m, 1H), 5.19 (m, 1H), 4.47 (m, 5H), 3.86 (m, 1H), 3.52 (m, 3H), 2.99 (s, 3H), 2.47 (m, 1H), 2.33 (m, 6H), 2.25 (m, 1H). MS: calc'd 685.2; found 686.3 (ESI+).
5’,3’-O-bis-p-toluoyl-2’-deoxyribose-1’-α-acetal R
1H data (400.115 Hz), δ: 7.83 (m, 4H), 7.45 (dd, 2H, J = 8.4 Hz, J = 34 Hz), 7.28 (dd, 2H, J = 8.4 Hz, J = 21.6 Hz), 7.16 (m, 4H), 7.00 (m, 2H), 6.72 (d, 1H, J = 16.4 Hz), 6.62 (d, 1H, J = 8.8 Hz), 6.47 (m, 1H), 6.26 (d, 1H, J = 12.4 Hz), 5.48 (m, 1H), 5.33 (m, 1H), 5.22 (m, 1H), 4.53 (m, 1H), 4.45 (m, 2H), 3.85 (m, 1H), 3.53 (m, 2H), 2.93 (s, 3H), 2.45 (m, 2H), 2.32 (m, 6H). MS: calc’d 630.3; found 631.3 (ESI+).
Deoxyriboside J
1H-NMR data (400.115 Hz), δ: 8.16 (dd, 2H, J = 9.6, 2.7 Hz), 7.74 (d, 2H, J = 9.0 Hz), 7.47 (d, 2H, J = 7.6 Hz), 7.21 (d, 1H, J = 16.3 Hz), 6.94 (d, 1H, 16.1 Hz), 6.74 (m, 2H), 5.23 (m, 1H), 4.48 (bm, 1H), 4.1 (bm, 1H), 4.08 (m, 1H), 3.98 (m, 1H), 3.72 (m, 6H), 2.95 (m, 3H), 2.35 (m, 1H), 2.18 (m, 1H). 13C (100.618 Hz): 150.2, 145.9, 134.7, 129.3, 127.0, 124.7, 121.6, 112.5, 104.3, 87.7, 86.6, 71.5, 65.0, 63.4, 61.9, 52.1, 41.9, 39.2. MS: calc'd 414.2; found 415.2 (ESI+).
Deoxyriboside K
1H-NMR data (400.115 Hz), δ: 7.41 (m, 3H), 6.72 (m, 2H), 6.53 (s, 1H), 6.41 (s, 1H), 6.41 (d, 1H, J = 15.7 Hz), 5.19 (d, 1H, J = 4.6 Hz), 4.47 (bs, 1H), 4.10 (bs, 1H), 4.05 (m, 1H), 3.98 (m, 2H), 3.05 (s, 3H), 2.36 (s, 3H) 2.27 (m, 1H), 2.14 (m, 1H). 13C (100.618 Hz): 162.0, 160.8, 156.8, 151.1, 138.9, 130.0, 122.7, 115.9, 112.8, 112.1, 106.4, 105.7, 104.9, 87.8, 72.9, 65.0, 63.2, 57.4, 52.0, 41.9, 39.0, 20.2. MS: calc'd 449.2; found 450.2 (ESI+).
Deoxyriboside R
1H-NMR data (400.115 Hz), δ: 7.59 (dd, 2H, J = 0.8 Hz, J = 8.5), 7.51 (dd, 2H, J = 1.7 Hz, J = 8.4 Hz), 7.42 (dd, 2H, J = 1.1 Hz, J = 8.7 Hz), 7.15 (d, 1H, J = 16.3 Hz), 6.88 (d, 1H, J = 16.3 Hz), 6.73 (m, 2H), 5.22 (m, 1H), 4.52 (bm, 1H), 4.10 (bs, 1H), 4.08 (m, 1H), 3.97 (m, 1H), 3.72 (m, 6H), 3.02 (d, 3H, J = 5.4 Hz), 2.22 (m, 1H), 2.07 (m, 1H). 13C (100.618 Hz): 149.7, 143.0, 128.5, 126.5, 125.0, 124.8, 122.4, 119.6, 112.6, 109.5, 104.8, 87.6, 77.5, 72.3, 63.6, 52.6, 42.9, 39.2. MS: calc'd 394.2; found 395.2 (ESI+).
5’-Dimethoxytrityl deoxyriboside J
H-NMR data (400.115 Hz), δ: 8.20 (d, 2H, J = 8.9 Hz), 7.58 (d, 2H, J = 8.8 Hz), 7.45 (d, 2H, J = 7.0 Hz), 7.36 (m, 10H), 7.21 (d, 1H, J = 16.2 Hz), 6.92 (d, 1H, J = 16.2 Hz), 6.82 (d, 4H, J = 8.5 Hz), 6.59 (d, 2H, J = 8.8 Hz), 5.13 (dd, 1H, J = 1.8 Hz, J = 5.2 Hz), 4.44 (m, 1H), 3.96 (m, 1H), 3.76 (s, 6H), 3.5 (m, 1H), 3.43 (m, 1H), 3.30 (dd, 1H, J = 5.2 Hz, J = 9.5 Hz), 2.88 (s, 3H), 2.21 (m, 1H), 2.07 (m, 1H), 1.81 (d, 1H, J = 4.3 Hz). 13C (100.618 Hz): 158.5, 149.6, 145.9, 145.1, 144.9, 136.0, 133.7, 130.1, 128.5, 128.2, 127.9, 126.9, 126.1, 124.2, 124.1, 121.4, 113.2, 111.9, 104.2, 86.2, 84.7, 73.4, 65.0, 64.9, 55.3, 52.1, 41.3, 38.0. MS: calc'd 716.3; found 716.9 (ESI+). HRMS: calc'd 716.3098; found 739.2995 (ESI+, w/ Na+).
5’-Dimethoxytrityl deoxyriboside K
1H-NMR data (400.115 Hz), δ: 7.34 (m, 4H), 7.19 (m, 5H), 7.09 (m, 4H), 6.73 (m, 4H), 6.64 (m, 1H), 6.48 (m, 1H), 6.39 (m, 1H), 6.35 (d, 1H, J = 15.6 Hz), 5.79 (dd, 1H, J = 2.4 Hz J = 12.8 Hz), 5.46 (s, 1H), 5.14 (d, 1H, J = 4.6 Hz), 5.04 (m, 1H), 4.32 (m, 1H), 4.05 (m, 2H), 3.89 (m, 2H), 3.69 (s, 6H), 3.46 (m, 3H), 3.03 (m, 1H), 2.98 (m, 3H), 2.81 (s, 1H). 13C (100.618 Hz): 162.0, 160.7, 158.8, 156.8, 151.0, 147.6, 145.0, 139.7, 138.8, 136.2, 136.1, 130.3, 130.0, 129.4, 128.3, 128.0, 127.3, 115.9, 113.4, 113.0, 112.3, 106.4, 105.7, 105.0, 87.8, 87.1, 86.3, 73.5, 64.9, 64.2, 55.5, 52.2, 41.5, 39.1. MS: calc'd 751.32; found 751.7 (ESI−). HRMS: calc'd 774.3159; found 774.3155 (ESI+, w/ Na+).
5’-Dimethoxytrityl deoxyriboside R
1H-NMR data (400.115 Hz), δ: 7.58 (m, 1H), 7.50 (m, 2H), 7.41 (m, 4H), 7.29 (m, 5H), 7.18 (m, 4H), 7.10 (m, 1H), 6.83 (m, 4H), 6.73 (m, 1H), 6.59 (m, 1H), 5.23 (m, 1H), 4.13 (m, 2H), 3.96 (m, 1H), 3.80 (s, 6H), 3.12 (d, 2H, J = 4.4 Hz), 3.01 (s, 3H), 2.79 (s, 3H), 2.20 (m, 1H), 1.98 (m, 1H). 13C (100.618 Hz): 158.6, 158.4, 149.4, 147.3, 144.7, 139.4, 135.9, 135.8, 133.2, 132.5, 132.3, 131.9, 130.2, 130.0, 129.4, 129.1, 128.3, 128.1, 127.8, 127.7, 127.0, 126.7, 126.2, 122.1, 113.1, 112.0, 111.4, 104.7, 92.4, 86.9, 81.4, 73.3, 64.7, 63.9, 55.2, 52.0, 46.2, 41.2, 38.7, 11.7. MS: calc'd 696.3; found 697.4 (ESI+). HRMS: calc'd 696.3199; found 719.3097 (ESI+, w/ Na+).
ODF library construction and screening
The synthesis of the tetramer library was carried out as described before and was accomplished by a split and pool synthetic strategy using the monomers Y,E,B,S,D,K,R,J (Fig. 1).30 The assembly of the polyfluorophore chain was accomplished by automated DNA synthesis (1 μmol scale) on ABI 394 DNA synthesizer through Phosphoramidite chemistry. The fluorophore sequences of each library member were recorded by the binary encoding strategy with molecular tags. The tag synthesis, tagging and decoding procedure was done according to the published procedure by Still.31
The polyfluorophore library was screened under an epifluorescence microscope (Nikon Eclipse E800 equipped with 4x objective, excitation 340−380nm; emission >400 nm). Fluorescence images were taken using a Spot RT digital camera and Spot Advanced Imaging software. Selected beads were picked up with a flame-pulled pipet and transferred into a capillary tube for decoding. The sequences decoded are listed in Table S1 according to their approximate colors and intensities.
Oligodeoxyfluoroside Synthesis
Oligodeoxynucleotides were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer on a 1 or 0.5 μmole scale and possessed a 3’-phosphate group. Coupling employed standard β-cyanoethyl phosphoramidite chemistry, but with extended coupling time (600 s) for nonnatural nucleotides. Coupling efficiencies exceeded 60%. Oligomers were deprotected in 0.05 M potassium carbonate in methanol. Purification was carried out utilizing a Shimadzu 10 Series HPLC with an Alltec C4 column with acetonitrile and water as eluents.
Optical methods
Absorbance spectra were obtained on a Cary 100 Bio UV-vis spectrometer. Fluorescence studies were performed on a Jobin Yvon-Spex Fluorolog 3 spectrometer. Emission lifetime measurements were carried out on a PTI Easylife using either 340, 385 or 450 nm LED. Methyl viologen dichloride hydrate (98%) was purchased from Sigma-Aldrich. Solutions of 1−23 were prepared to an optical density of 0.05 or less in phosphate buffered saline (pH = 7.2) in order to minimize inner filter effects. Perylene in cyclohexane was used as a reference for quantum yields.27 Quantum yields were calculated according to the equation:
where Φ is the quantum yield for photoemission, A is the magnitude of the absorbance at the excitation wavelength, E is the integrated emission intensity, I is the intensity of the excitation wavelength and n is the refractive index of the solvent; the subscripts R and S indicate values for the reference and sample respectively.
Cell uptake studies
Cell Culture
HeLa CCF-2 cells (ATCC) were maintained in Dubelcco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicilin and 100 μg/ml streptomycin (all reagents from Invitrogen) in a humidified atmosphere at 37°C with 5% CO2.
Cellular uptake and colocalization
Cells were plated in chambered coverglass (0.5 ml medium/chamber) and allowed to reach 60% confluence in 1−2 days. The growth medium was removed and new growth medium containing 10 μM of SSYB or 5 μM SSSBEY was added to the cells and incubated for 30 min - 3 h at 37 °C under 5% CO2. Cells were then washed thrice with PBS. Incubation with wheat germ agglutinin Alexa Fluor 633 conjugate at 5 ug/ml for 10 min was then carried out. Cells were rinsed three times with PBS and fresh media was added before imaging.
Confocal imaging
Live HeLa cells were visualized directly in chambered coverglass using a Leica SP2 AOBS confocal laser scanning microscope (Leica Microsystems, Heidelberg) viewed through a 63X emulsion objective. A blue diode laser (405nm) was used to excite the ODFs while the HeNe 633 nm laser was used for Alexa Fluor 633. Optical sections through the z-dimension were acquired in step sizes of ca. 0.326 μm. Image acquisition was performed at the Cell Sciences Imaging Facility of Beckman Center for Molecular and Genetic Medicine, Stanford University Medical Center, Stanford CA.
Zebrafish embryo labeling methods
Zebrafish were maintained at 28 °C and embryos were raised in E3 buffer. Solutions of 2, 7, 11, and 14 were prepared to 5 μM in E3. Embryos were removed from stock E3 buffer at 24 hours old via suction and distributed in groups of 4−6 on a 96-well microplate with a minimum volume of transfer solution. To each well was added 200−300 μL of polyfluor doped E3 solution and the embryos were incubated for 24 hours. For imaging, embryos and larvae were collected, rinsed with fresh E3 solution, collected and placed in fresh E3 solution on a microscope slide. No tricaine or agarose was used to sedate or stabilize the embryos or larvae. Experiments were carried out in accordance with the Administrative Panel on Laboratory Animal regulations and approvals.
Results
Oligodeoxyfluoroside design for multispectral imaging
Our goal for this study was to evaluate whether a set of ODF dyes could be identified that could be excited by long-wavelength UV light and emit a range of wavelengths across the visible spectrum. Previous ODF designs had incorporated fluorescent deoxyriboside monomers primarily emitting at blue and green wavelengths.18,30 Examples include pyrene (Y), benzopyrene (B), perylene (E), and dimethylaminostilbene (D) deoxyriboside monomers (Fig. 1). To better cover the full visible spectrum in the present study, we chose three new monomer chromophore designs (styrene/stilbene derivatives (J,K,R in Fig. 1)) that were expected to emit at 500−620 nm, thus more effectively covering yellow to orange-red emission wavelengths. Most, although not all, of these monomers can absorb light efficiently at the intended excitation wavelengths (340−380 nm). We considered the possibility that chromophores could be excited both by direct absorption and also by transfer of energy via FRET, excimer, exciplex, and other electronic interactions.
The ODF design is based on short, DNA-like phosphate diester oligomers of fluorescent monomers such as the above (see Fig. 2). The flat aromatic chromophores have the potential to interact both physically (by stacking)19 and electronically, thus yielding new emissive properties that the monomers alone do not have. In the present study we limited the length to tetramers, which is short enough to allow for high synthesis yields, yet long enough to allow for substantial degrees of emissive complexity in thousands of combinations.30 The library also contained a non-fluorescent spacer (S, Fig. 1) as one of the monomers so that ODFs containing one, two or three chromophores could be evaluated along with tetrameric cases.
Figure 2.
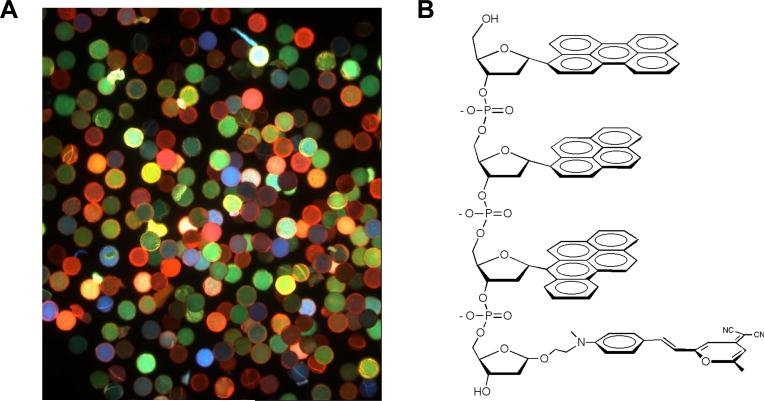
Multispectral oligodeoxyfluorosides. (A) Image from 4096-member tetramer ODF library on PEG-polystyrene beads, taken with 340−380 nm excitation. Library contains all possible combinations of the eight monomers in Fig. 1. (B) Structure of a typical ODF tetramer; this sequence is EYBK (listed in 5’->3’ direction), a red-orange ODF tetramer identified from this library.
Monomer and library synthesis
The monomer deoxyribosides with pyrene, perylene, dimethylaminostilbene, and benzopyrene substituted at C-1 (Figure 1) were prepared as described previously.18,20 The three new stilbene chromophores were prepared as deoxyribosides by formation of the acetal derivatives at C-1, using hydroxyethyl-substituted dyes. All seven fluorophore deoxyribosides were prepared as 5’-dimethoxytrityl, 3’-cyanoethylphosphoramidite derivatives, so as to be compatible with standard oligonucleotide solid-phase synthesis methods. Together with the tetrahydrofuran spacer moiety (S), there were eight monomers to be incorporated into the tetramer library. This yields 4096 possible ODF sequences in all combinations.
The tetramer ODF library was constructed on 60 μm PEG-polystyrene beads using standard split-and-mix methods. Thus each bead contained one tetramer sequence. Overall coverage was approximately 35-fold; i.e., each sequence was represented ca. 35 times in the full library, based on library size (4096 members) and total number of beads. The monomers incorporated were encoded by Still methodology,31 so that sequences could ultimately be decoded by electron capture gas chromatographic analysis of tags liberated from each individual bead that was selected in the screening. Coupling yields for all monomers exceeded 60%, and averaged 80%, based on standard dimethoxytrityl cation monitoring on the DNA synthesizer.
Screening results
Screening of the library was carried out with dry beads in air, using an epifluorescence microscope to image ca. 200 beads at a time in a field of view. For excitation we used a filter set with an excitation window of 340−380 nm and for emission, a long-pass filter (cutoff 400 nm). This allowed us to visualize the entire visible spectrum by eye, without changing filters. A typical field of view is shown in Fig. 2. Inspection revealed a broad range of colors; dyes were typically well-saturated and appeared to be dominated by one color in the majority of cases, with white or whitish hues in the minority. Intensities of emission varied greatly, from nonemissive cases to apparently bright cases.
We picked and sequenced ca. 80 beads of varied hues, focusing chiefly on saturated colors and the brightest intensities. A full listing of selected sequences is given in the Supporting Information (SI) file. For further investigation in solution, we chose 23 cases to re-synthesize (Table 1). Twenty of these contained two, three, or four monomer chromophores. For comparison, we also examined the three single-monomer cases containing pyrene, perylene, and benzopyrene. All of these (except the monomers) are expected to be water soluble owing to the anionic phosphate groups between the monomers; however, to ensure water solubility and minimize aggregation tendency, we added terminal spacer residues (S) to all, thus adding charges without increasing the numbers of hydrophobic aromatic hydrocarbons. Resynthesized ODFs were characterized by mass spectrometry (see SI) to confirm that all monomers were coupling correctly and were retained intact in the final purified ODF. The twenty-three fluorophores selected for further study contained various combinations of only four chromophore monomers of the original seven: pyrene (Y, Figure 1), perylene (E), benzopyrene (B), and 4-(dicyanomethylene)-2-methyl-6-(p-dimethylaminostyryl)-4H-pyran (commonly abbreviated as DCM) (K); spectral data for the monomeric fluorophores are shown in Figs. 4A and 5A (for Y, E, and B), and in the Supporting Information file (for K). Despite this small number of monomer components, the 23 oligomeric compounds display a broad spectrum of visible colors (Fig. 3). Sequences are listed as if they were DNA, in 5’-to-3’ direction, using single letter abbreviations. For simplicity, the terminal spacers (S) are omitted in the discussions below, but they are shown in the Table.
Table 1.
Optical data for ODF fluorophores in this study.
| Entry | Sequence | λmax, abs | ε | λmax, em | Φa | τ, ns |
|---|---|---|---|---|---|---|
| 1 | 5’-SY-3’ | 327, 343 | 46,000 | 376, 396 | 0.37 | 177±4.5 |
| 2 | 5’-SB-3’ | 375, 395 | 28,000 | 427 | 0.91 | 10.4±.7 |
| 3 | 5’-SE-3’ | 415, 441 | 39,000 | 412 | 0.86 | 5.2±.1 |
| 4 | 5’-SSBY-3 | 344, 380, 400 | 49,000 | 497 | 0.20 | 10.2±.8, 45% 101.5±2.7, 55% |
| 5 | 5’-SSSYYY-3’ | 328, 344 | 67,000 | 491 | 0.12 | 114±9.3 |
| 6 | 5’-SSSBEY-3’ | 346, 396, 450 | 50,000 | 376, 492 | 0.42 | 6.1±.7, 58% 26.9±2.8, 42% |
| 7 | 5’-SSEY-3’ | 346, 448 | 47,000 | 464, 490 | 0.65 | 5.1±.9, 46% 30.8±2.3, 54% |
| 8 | 5’-SSSBYE-3’ | 345, 398, 447 | 50,000 | 377, 496 | 0.18 | 6.8±.1, 63% 47.7±.1, 37% |
| 9 | 5’-SSSEBY-3’ | 344, 397, 447 | 50,000 | 520 | 0.29 | 7.6±.1, 56% 41.6±., 44% |
| 10 | 5’-SSEB-3’ | 396, 448 | 39,000 | 412, 519 | 0.26 | 5.1±.9, 46% 30.8±2.3, 54% |
| 11 | 5’-SSBB-3’ | 380 | 48,000 | 412, 509 | 0.31 | 8.9 ± .4, 71% 61.4 ± 4.8, 29% |
| 12 | 5’-SSSEYK-3’ | 326, 425, 450 | 61,000 | 488 | 0.17 | 3.4±.9, 91% 34.6±7.8, 9% |
| 13 | 5’-SSEE-3’ | 443, 420 | 43,000 | 449, 478, 560 | 0.06 | 3.2±.6, 54% 20.6±2.8, 46% |
| 14 | 5’-SSSBEK-3’ | 397, 443 | 53,000 | 448, 631 | 0.07 | 4.2± .1 |
| 15 | 5’-SSSKYY-3’ | 330, 446, 477 | 89,000 | 482, 602 | 0.05 | 7.1± .5 |
| 16 | 5’-SSSEKB-3’ | 401, 444 | 53,000 | 414, 447, 550 (bs)b | 0.18 | 4.7±.1, 86% 52.9±9.0, 14% |
| 17 | 5’-SSSYKY-3’ | 345, 490 | 81,000 | 380, 487,624 | 0.06 | 1.7 ± 0.1, 92.% 14.4 ± 1.4, 8% |
| 18 | 5’-SSSBKBK-3’ | 381, 444 (b)b | 71,000 | 415, 625 | 0.09 | 1.8 ±.2, 73% 6.5±1.3, 27% |
| 19 | 5’-SSSYKBK-3’ | 379 | 92,000 | 414, 604 | 0.04 | 4.1 ± 0.3, 83% 16.7 ± 1.5, 17% |
| 20 | 5’-SSSEYBK-3’ | 346, 398, 448 | 75,000 | 414, 624 | 0.028 | 1.7±.1, 82% 5.6±1.0, 18% |
| 21 | 5’-SSBK-3’ | 396, 480 | 40,000 | 412, 633 | 0.10 | 2.3±.1, 91% 10.7±.1, 9% |
| 22 | 5’-SSSYKEB-3’ | 326, 399, 450 | 72,000 | 412, 495, 626 | 0.12 | 3.0±.1, 77% 11.2±.1, 23% |
| 23 | 5’-SSSYKYK-3’ | 346, 477 | 110,000 | 380, 482, 629 | 0.09 | 2.3±.1, 96% 45.3±1.7, 4% |
Perylene as a reference; error ± 10%.
(b = broad, bs = broad shoulder).
Figure 4.
Normalized absorption spectra of 23 ODFs from Table 1 in water. (A) Sequences 1−5; (B) sequences 6−10; (C) sequences 11−15; (D) sequences 16−20; (E) sequences 21−23.
Figure 5.
Normalized emission spectra of 23 ODFs from Table 1 in water, with 354 nm excitation. (A) Sequences 1−5; (B) sequences 6−10; (C) sequences 11−15; (D) sequences 16−20; (E) sequences 21−23.
Figure 3.
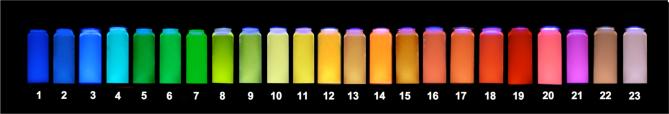
Combined photograph of 23 multispectral ODFs identified from the library (listed in Table 1), dissolved in phosphate buffer. The ODF solutions were excited by a UV transilluminator (λex = 354 nm) and concentrations were adjusted to show relative colors at similar brightness. These ODFs contain combinations of only four different chromophores (Y,E,B,K).
Optical properties of ODFs
Optical data were measured for the 23 ODFs in water. Table 1 lists absorption and emission maxima, extinction coefficients, fluorescence quantum yields, and emission lifetimes for the compounds. The corresponding data for the monomer components are also listed in the Table and SI. Absorption and emission spectra of the ODFs are shown in Figures 4 and 5, respectively. Overall, the data show a broad range of absorption and emission properties. Apparent colors of the ODFs in water over a UV transilluminator range from violet to red (Fig. 3). Quantum yields range from 0.03 to 0.91, and emission lifetimes vary from 2 to 180 ns. In general, the absorption spectra resemble the expected combination of the monomer chromophore components, although line broadening is evident in many cases (Fig. 4), consistent with ground-state stacking between the aromatic hydrocarbons. In contrast to this, most of the emission spectra of the ODFs (Fig. 5) are quite different than the simple combination of components, which is suggestive of extensive excited-state interactions among the chromophores.
Evidence of electronic interactions within ODFs
Closer examination of emission spectra for a number of ODF cases reveals emissive properties that vary not only with the compositions of chromophore components, but also with the sequence in which they occur. For example, comparisons of dimers containing pyrene, benzopyrene and perylene with the monomers shows loss of the characteristic structured violet/blue emission bands of the parent hydrocarbons, and appearance in all cases of long-wavelength broadened peaks that are consistent with excimer and exciplex emission.19,32 These differences are evident in sequences 4,5,7,8 as compared with component dyes in 1−3 (see Figs. 5A,B). The apparent excimer- and exciplex-forming cases include BY, EY, EB, and BB, which have emission maxima from 490−520 with very little of the monomer emission peaks evident. These dimers have high quantum yields of 0.20−0.65 and elevated extinction coefficients of 39,000−49,000.
Also unusual are “anagram” cases wherein the chromophore composition is the same, but the sequence is different. For example, the ODF isomers KYY (seq. 15 in Table 1) and YKY (17) show very similar absorption spectra but distinctly different emissions (Fig. 6A). The KYY case shows two emission peaks at 482, 602 nm. In contrast, the emission of the YKY fluorophore is dominated by a 380-nm band; two longer-wavelength bands are also evident, but at substantially different wavelengths (487, 624 nm) than the former anagram. A second comparison can be made from two tetramer anagrams in the set: the sequences YKEB (compound 22) and EYBK (20). The former is a nearly white-colored fluorophore with two main emission bands at 495 and 626 nm (Fig. 6C). In contrast to this, the latter appears pink in hue, and its emission is dominated by a single band at 624 nm. Also noteworthy is the fact that both ODF spectra differ markedly from the combination of the component fluorophores (Fig. 6D). Thus these cases appear to be highly sensitive to the interaction and ordering of neighboring chromophores.24 Interestingly, not all cases display a sequence dependence that is this strong; for example, the anagrams BEY, BYE, EBY (6,8,9 respectively) all have similar (although not identical) emission spectra (see Fig. 5B).
Figure 6.
Fluorescence spectra of ODF sequence anagrams, showing differences in emission with varied monomer order. (A) Comparison of KYY and YKY emission spectra; (B) spectra of Y and K separately. (C) Comparison of YKEB and EYBK emission spectra; (D) spectra of E, Y, B, K separately. Excitation at 354 nm in water. The K spectrum is for the free nucleoside in methanol. Spectra are shown with their approximate colors.
Preliminary studies of cell uptake of ODFs
The spectral properties of many of the ODFs in Table 1 suggest that they might be useful in biological applications; however, it is unclear how they might interact with cellular systems. These ODFs carry 2−6 negative charges, but also possess substantial amounts of hydrophobic structure; thus they may be amphiphilic in character. To begin to evaluate how such dyes might interact with human cells, we chose two cases (SSYB and SSSBEY), and incubated them with HeLa cells. We then evaluated their interactions with the cells by laser confocal microscopy. Data for one case (SSYB) are shown in Fig. 7; the second (SSSBEY, a yellow dye) showed similar results, and data are given in the SI file. In the experiments, we used a commercial membrane-labeling dye as a reference for cellular localization. Simple incubation for 3 h showed that the green SSYB ODF was taken up into the cytoplasm. It was not apparently associated with the outer cell membrane, and it did not significantly enter the nucleus. The fact that the color remained unchanged suggested that the compound was biostable over at least several hours. Thus it apparently was not degraded by intracellular nucleases, which would have separated the component chromophores and changed the emission.
Figure 7.
Laser confocal microscopic images of HeLa cells after incubation with SSYB ODF dye (green). The cell membranes were marked with Alexa Fluor 633-wheat germ agglutinin (red). (A) Image of multiple cells, showing apparent cytoplasmic localization of the ODF. Nuclei are unstained and appear dark at the center of each cell. (B) Successive images of xy focal plane moved in the z dimension through cells, confirming cytoplasmic location of green ODF dye. Focal plane was moved ca. 1 μM per frame. Similar results were obtained with the SSSBEY ODF, a yellow dye (see SI).
Multispectral labeling of zebrafish embryos
One possible application of multispectral dyes is in dynamic systems, where tracking of multiple moving species is desired. Living organisms offer such a system, and so we tested a set of four distinctly colored ODFs selected from the 23-member set for possible labeling of vertebrate embryos. The ODFs chosen for the experiments were SB (2, blue), SSEY (7, cyan), SSBB (11, yellow), and SSSYYK (15, orange-red). The dyes were separately incubated at 5 μM with fertilized zebrafish (Danio rerio) eggs, and the resulting embryos were combined and imaged at 24 and 48 h. No toxicity as a result of the dye at 5 μM was noted, and most embryos were viable after 48 h whether or not they were incubated in the presence of one of the dyes. Images of the labeled embryos are shown in Fig. 8. The 24 h-old embryos showed association of the dyes with the egg outer membrane (chorion), with at least some penetration to the body of the developing fish visible with the blue and cyan dyes. At 48 h the embryos showed similar labeling; however, in fish that had hatched and were free-swimming, the ODFs (again, in the blue and cyan cases at least) showed apparent penetration into tissues, with internal structures of the head visible. As in the human cell labeling, no apparent degradation of the dyes was observed. Importantly, excitation of all four colors was done with UV excitation (340−380 nm), and a simple long-pass UV emission filter allowed observation of the moving fish by eye under the microscope, without switching of filters. The images shown in Fig. 8 are real-color images, made without reconstruction of separate false-colored R,G,B images as is typical of biological multicolor labeling with commercial organic dyes.
Figure 8.
The use of four ODF dyes in simultaneous observation of differently labeled live zebrafish embryos. The ODFs are 2 (blue); 7 (cyan); 11 (yellow); and 14 (red), and were separately incubated with embryos at 5 uM. (A) Embryos at 24 h post-fertilization. (B) Embryos at 48 h post-fertilization. Note staining of hatched, free-swimming fish as well as chorion of unhatched embryos. These are real images taken with a single long-pass filter (excitation 354 nm).
Discussion
The current data show that the set of 23 ODF dyes exhibits a broad range of emission properties. An earlier study described how rationally varied combinations of two monomers could result in up to 6 visibly different emission hues;24 however, quantum yields of ODFs that were not green or blue were low in that limited set. That motivated us in the present work to prepare new monomers (J,K,R) that have stronger emission at wavelengths beyond 500 nm, and to include all monomers in a larger library of several thousand members. The present results show that screening such a larger library resulted in ODFs with considerably more favorable spectral coverage and improved quantum yields.
The data generated for the 23 ODFs in Table 1 show that they exhibit a wide range of emissive behavior, with apparent colors from violet to red across the visible spectrum. However, unlike commercial dye sets (such as the common Alexa fluors™), the ODFs can all be excited simultaneously, with one filter set. This allows for increased simplicity of use when multiple species are to be imaged simultaneously. With current 3- and 4-color images in biology, the standard methods require the experimenter to make multiple separate images, switching between different excitation emission filter sets. These images are false-colored and then overlaid digitally to obtain a reconstructed multicolor image. This fact not only requires expensive equipment, but also prevents the experimenter from observing the multiple species in real time by eye. The requirement for switching excitation light and emission filters also prevents the imaging of rapidly moving systems in more than one color. In contrast to this, the current experiments have demonstrated the imaging of moving fish in four colors.
Although the current study reports data on 23 different ODF fluorophores, it is unlikely that a set this large would routinely be used simultaneously. First, a number of the dyes overlap spectrally, and thus would be difficult to distinguish from one another without more detailed spectral analysis. By eye, one can distinguish at least 15 different hues from this set (Fig. 3), which is a more realistic limit for multicolor application in routine experiments. It should also be noted that because of the electronic complexity in the ODFs, many of them exhibit emission lines that are less narrow than some common commercial dyes. The peak widths of the 23-member set range from ca. 40 nm to as much as ca. 140 nm at half-height, whereas common fluorescein dyes have widths of ca. 50 nm. Moreover, some ODFs have more than one emission band, which can also lead to overlap. Interestingly, however, the electronic interactions in the ODFs often simplify the emission as compared with the monomer components: for example, the EYBK sequence has essentially a single dominant emission band at 624 nm, whereas the monomer components show bands at ca. 380, 400, 410, 430, and 615 nm. Such results suggest that this (and many other) ODFs act as a single electronic unit rather than as a simple combination of dyes. Overall, the limits on multianalyte imaging with ODF dyes would depend on whether the species overlap in space, and whether equipment or software exists (in a given experiment) that can deconvolute the spectra based on variations in their emission peaks.
The current ODF dye set also has the advantage over common organic dyes of large Stokes shifts, which range from 30 to over 230 nm. Thus the ODFs are not expected to suffer from interference by the exciting light, whereas common organic dyes often have small Stokes shifts of 10−30 nm, and so careful filtering is necessary. In this respect, ODF dyes have some of the useful properties of semiconductor quantum dots (QDs), which can also be excited by long-wavelength UV light for multispectral analysis.10-12 QDs are commercially available in several emission colors, based on different sizes. One advantage of QDs over ODF dyes is the high photostability offered by the inorganic nanoparticles. On the other hand, ODF fluorophores offer a number of advantages over QDs. First, they are discrete, soluble organic molecules, and can easily be conjugated to other species (such as nucleic acids, peptides, and small molecules) using commercial and/or published methods that have been developed for synthetic DNA.33 Second, they appear (at least in some cases) to be cell permeable as well.
Recent studies have reported the synthesis of libraries of small-molecule fluorophores containing dozens to hundreds of different dyes.34,35 Such dyes have been evaluated individually for their emission properties. One useful feature of the current oligomeric fluorophores, by contrast, is that they can be made in large numbers of combinations with great electronic complexity, and libraries can be screened rapidly not only for simple emission properties (such as in the current study) but also for sensing molecular species and physical properties in solution.30 We have described libraries containing over 14,000 different fluorophores using this approach,30 and have found sensors of light exposure, of organic species, and of metal ions.34 This combinatorial nature increases the ease of synthesis as well. For example, the current 23 fluorophores are composed of only four different emissive monomers. Thus all 23 could be assembled in one day on a synthesizer, and are all constructed on solid support using automated chemistry.
Studies by Frechet,37,38 Müllen,39,40 and others41,42 have described large dendrimeric molecules that carry multiple chromophores for gathering light, and in some cases, a second chromophore that accepts this energy. These classes of molecules could potentially offer some of the useful properties of ODFs. However, their construction is considerably more complex than is the automated synthesis of ODFs. Moreover, methods for making discrete bioconjugates of dendrimers43,44 are limited compared with the many existing methods of conjugating DNA.34 Finally, the linear iterative approach that we use to assemble our ODFs leads to very straightforward application of combinatorial methods, which have not generally been applied to dendrimeric molecules.
A number of other laboratories have investigated the use of multiple chromophores on a DNA scaffold. For example, researchers have studied the concept of multi-FRET interactions by placing two or more dyes conjugated to DNA bases at intervals along a DNA strand.45-50 However, the current approach produces considerably greater electronic complexity and orders-of-magnitude greater numbers of distinct fluorescent molecules, and achieves this in much smaller structures. In addition to this approach of conjugating chromophores to otherwise standard DNA, a number of laboratories have begun recently to adopt the current, more direct approach of incorporating multiple fluorescent DNA bases or related aromatic species into DNA.51-57 Their fluorescence properties and applications are still under early investigation, but the current data suggest that such approaches can yield useful properties.
Our very early data with two ODFs demonstrate human cell permeability and cytoplasmic localization. Moreover, staining of vertebrate tissue also appears possible. However, it is unclear how this membrane permeability depends on ODF composition, length, and charge. Further studies will be needed to address this issue before such fluorophores are broadly applied in intracellular applications.
In conclusion, the current study shows that it is possible to identify a broad set of multispectral oligomeric, water-soluble fluorophores that can be imaged simultaneously. The ease of synthesis of a larger set of widely varied dyes from a small set of monomers makes them attractive as candidates for more general use. Moreover, the iterative synthesis makes their assembly into combinatorial libraries simple, which then enables rapid screening for varied properties. Indeed, early studies have also shown that not only static fluorophores, but also color-changing sensors may be developed based on such a multichromophore scaffold.30 Future studies will be directed to exploring the use of ODF fluorophores to a range of chemical and biological problems.
Supplementary Material
Acknowledgement
We thank the National Institutes of Health (GM067201) for support. J.N.W. acknowledges an NIH postdoctoral fellowship, and Y.N.T acknowledges an A*STAR graduate fellowship.
Footnotes
Supporting Information Available: Full listing of ODF sequences picked and decoded from library; mass spectrometry data for all ODF oligomers; cell uptake data for additional ODF sequence.
Literature Cited
- 1.Marks KM, Nolan GP. Nat. Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 2.Weissleder R. Nat. Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 3.Wouters FS, Verveer PJ, Bastiaens PI. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 4.So PT, Dong CY, Masters BR, Berland KM. Annu. Rev. Biomed. Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 5.Nath J, Johnson KL. Biotech. Histochem. 2000;75:54–78. doi: 10.3109/10520290009064150. [DOI] [PubMed] [Google Scholar]
- 6.Asseline U. Curr. Org. Chem. 2006;10:491–518. [Google Scholar]
- 7.Ntziachristos V. Annu. Rev. Biomed. Eng. 2006;8:1–33. doi: 10.1146/annurev.bioeng.8.061505.095831. [DOI] [PubMed] [Google Scholar]
- 8.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 9.Steyer JA, Almers W. Nat. Rev. Mol. Cell. Biol. 2001;2:268–275. doi: 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- 10.Alivisatos P. Pure Appl. Chem. 2000;72:3–9. [Google Scholar]
- 11.Empedocles S, Bawendi M. Acc. Chem. Res. 1999;32:389–396. [Google Scholar]
- 12.Du W, Wang Y, Luo Q, Liu BF. Anal. Bioanal. Chem. 2006;386:444–457. doi: 10.1007/s00216-006-0541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller JE, Zugates GT, Ferreira LS, Ow HS, Nguyen NN, Wiesner UB, Langer RS. Biomaterials. 2008;10:1526–1532. doi: 10.1016/j.biomaterials.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Empedocles S, Bawendi M. Acc. Chem. Res. 1999;32:389–396. [Google Scholar]
- 15.Cady NC, Strickland AD, Batt CA. Mol. Cell. Probes. 2007;2:116–124. doi: 10.1016/j.mcp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Ju J, Ruan C, Fuller CW, Glazer AN, Mathies RA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4347–4351. doi: 10.1073/pnas.92.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao GS, Thoresen LH, Kim TG, Haaland WC, Gao F, Topp MR, Hochstrasser RM, Metzker ML, Burgess K. Chemistry. 2006;30:7816–7826. doi: 10.1002/chem.200600197. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Strassler C, Tahmassebi D, Kool ET. J. Am. Chem. Soc. 2002;124:11590–11591. doi: 10.1021/ja027197a. [DOI] [PubMed] [Google Scholar]
- 19.Cuppoletti A, Cho Y, Park JS, Strässler S, Kool ET. Bioconj. Chem. 2005;16:528–534. doi: 10.1021/bc0497766. [DOI] [PubMed] [Google Scholar]
- 20.Ren RX-F, Chaudhuri NC, Paris PL, Rumney S, IV, Kool ET. J. Am. Chem. Soc. 1996;118:7671–7678. doi: 10.1021/ja9612763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strässler C, Davis NE, Kool ET. Helv. Chim. Acta. 1999;82:2160–2171. doi: 10.1002/(sici)1522-2675(19991215)82:12<2160::aid-hlca2160>3.0.co;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson JN, Cho Y, Tan S, Cuppoletti A, Kool ET. ChemBioChem. 2008;9:279–285. doi: 10.1002/cbic.200700381. [DOI] [PubMed] [Google Scholar]
- 23.Wilson JN, Teo YN, Kool ET. J. Am. Chem. Soc. 2007;129:15426–15427. doi: 10.1021/ja075968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson JN, Gao J, Kool ET. Tetrahedron. 2007;63:3427–3433. doi: 10.1016/j.tet.2006.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a Crespo-Hernández CE, Cohen B, Kohler B. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]; b Kwok WM, Ma C, Phillips DL. J. Am. Chem. Soc. 2006;128:11894–11905. doi: 10.1021/ja0622002. [DOI] [PubMed] [Google Scholar]; c Takaya T, Su C, de La Harpe K, Crespo-Hernández CE, Kohler B. Proc. Natl. Acad. Sci USA. 2008;105:10285–10290. doi: 10.1073/pnas.0802079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballet W, Picard I, Verbiest T, Persoons A, Samyn C. Macromol. Chem. Phys. 2004;205:13–18. [Google Scholar]
- 27.Sheeren G, Persoons A, Rondou P, Beylen MV, Samyn C. Eur. Polym. J. 1993;29:975–979. [Google Scholar]
- 28.Reynolds DD, Evans WL. J. Am. Chem. Soc. 1938;60:2559–2561. [Google Scholar]
- 29.Berlman IB. Handbook of fluorescence spectra of aromatic molecules. Academic Press; New York: 1965. [Google Scholar]
- 30.Gao J, Watanabe S, Kool ET. J. Am. Chem. Soc. 2004;126:12748–12749. doi: 10.1021/ja046910o. [DOI] [PubMed] [Google Scholar]
- 31.Ohlmeyer MHJ, Swanson M, Dillard LW, Reader JC, Asouline G, Kobayashi R, Wigler M, Still WC. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winnik FM. Chem. Rev. 1993;93:587–614. [Google Scholar]
- 33.Beaucage SL, Iyer RP. Tetrahedron. 1993;49:1925–1963. [Google Scholar]
- 34.Rosania GR, Lee JW, Ding L, Yoon H, Chang YT. J. Am. Chem. Soc. 2003;125:1130–1131. doi: 10.1021/ja027587x. [DOI] [PubMed] [Google Scholar]
- 35.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org. Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 36.Kim SJ, Kool ET. J. Am Chem. Soc. 2006;128:6164–6171. doi: 10.1021/ja0581806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adronov A, Robello DR, Frechet JMJ. J. Polym. Sci., Part A:Polym. Chem. 2001;39:1366–1373. [Google Scholar]
- 38.Adronov A, Frechet JMJ. Chem. Commun. (Cambridge, U.K.) 2000:1701–1710. [Google Scholar]
- 39.Weil T, Wiesler UM, Herrmann A, Bauer R, Hofkens J, De Schryver FC, Mullen K. J. Am. Chem. Soc. 2001;123:8101–8108. doi: 10.1021/ja010579g. [DOI] [PubMed] [Google Scholar]
- 40.Hofkens J, Maus M, Gensch T, Vosch T, Cotlet M, Kohn F, Herrmann A, Mullen K, De Schryver F. J. Am. Chem. Soc. 2000;122:9278–9288. [Google Scholar]
- 41.Wang S, Hong JW, Bazan GC. Org Lett. 2005;10:1907–1910. doi: 10.1021/ol047674p. [DOI] [PubMed] [Google Scholar]
- 42.Lijanova IV, Moggio I, Arias E, Vazquez-Garcia R, Martínez-García M. J. Nanosci. Nanotechnol. 2007;10:3607–3614. doi: 10.1166/jnn.2007.917. [DOI] [PubMed] [Google Scholar]
- 43.Arima H, Kihara F, Hirayama F, Uekama K. Bioconjug. Chem. 2001;12:476–84. doi: 10.1021/bc000111n. [DOI] [PubMed] [Google Scholar]
- 44.Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR., Jr. Chem. Commun. (Cambridge, U.K.) 2005:5739–5741. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- 45.Kawahara S, Uchimaru T, Murata S. Chem. Commun. (Cambridge, U.K.) 1999:563–564. [Google Scholar]
- 46.Tong AK, Jockusch S, Li Z, Zhu H, Akins DL, Turro NJ, Ju J. J. Am. Chem. Soc. 2001;123:12923–12924. doi: 10.1021/ja016904h. [DOI] [PubMed] [Google Scholar]
- 47.Tong AK, Li ZM, Jones GS, Russo JJ, Ju JY. Nat. Biotechnol. 2001;19:756. doi: 10.1038/90810. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Lu Y. Methods Mol Biol. 2006;335:257–271. doi: 10.1385/1-59745-069-3:257. [DOI] [PubMed] [Google Scholar]
- 49.Ohya Y, Yabuki K, Hashimoto M, Nakajima A, Ouchi T. Bioconj. Chem. 2003;6:1057–1066. doi: 10.1021/bc034028m. [DOI] [PubMed] [Google Scholar]
- 50.Benvin AL, Creeger Y, Fisher GW, Ballou B, Waggoner AS, Armitage BA. J. Am. Chem. Soc. 2007;129:2025–2034. doi: 10.1021/ja066354t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayer-Enthart E, Wagenknecht HA. Angew. Chem. Int. Ed. 2006;45:3372–3375. doi: 10.1002/anie.200504210. [DOI] [PubMed] [Google Scholar]
- 52.Wagenknecht HA. Ann. N. Y. Acad. Sci. 2007 December 20; [Online early Access]. DOI:10.1196/annals.1430.001. Published Online. [Google Scholar]
- 53.Samain F, Malinovskii VL, Langenegger SM, Häner R. Bioorg. Med. Chem. 2008;16:27–33. doi: 10.1016/j.bmc.2007.04.052. [DOI] [PubMed] [Google Scholar]
- 54.Aubert Y, Asseline U. Org. Biomol. Chem. 2004;23:3496–3503. doi: 10.1039/b410695d. [DOI] [PubMed] [Google Scholar]
- 55.Hrdlicka PJ, Babu BR, Sørensen MD, Harrit N, Wengel J. J Am Chem Soc. 2005;38:13293–13299. doi: 10.1021/ja052887a. [DOI] [PubMed] [Google Scholar]
- 56.Seo YJ, Lee IJ, Yi JW, Kim BH. Chem. Commun. (Cambridge, U.K.) 2007;27:2817–2819. doi: 10.1039/b707278c. [DOI] [PubMed] [Google Scholar]
- 57.Krueger AT, Kool ET. J. Am. Chem. Soc. 2008;130:3989–3999. doi: 10.1021/ja0782347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.