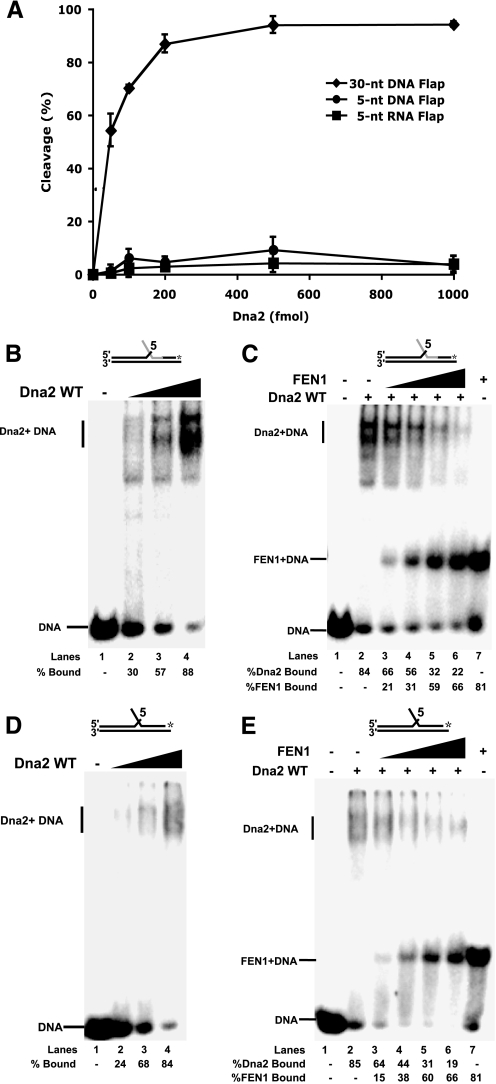
FIGURE 2.
FEN1 disengagement of Dna2 from short RNA and DNA flaps to which Dna2 binds but cannot cleave. A, Dna2 (50, 100, 200, 500, and 1000 fmol) was incubated with 5 fmol of a 5-nt RNA flap substrate (D1:U1:T1) (squares), a 5-nt DNA flap substrate (D2:U2:T2) (circles), or a 30-nt DNA flap substrate (D3:U2:T2) (diamonds). Cleavage activity was then measured by denaturing PAGE. B, gel shift analysis was used to measure Dna2 (0.2, 0.5, 1 pmol) binding activity on the 5-nt RNA flap (lanes 2–4). Lane 1 is the substrate alone control. C, Dna2 (1 pmol) was pre-bound to the 5-nt RNA flap substrate followed by the addition of FEN1 (5, 10, 25, and 50 fmol) (lanes 3–6). Thesampleswerethenanalyzedbygelshift.Lanes1and7arethesubstratealone and substrate plus FEN1 (50 fmol), respectively. WT, wild type. D, as described in B, except a 5-nt DNA flap was used. E, as described in C, except a 5-nt DNA flap substrate was used. A, points are an average of three experiments, and the bars indicate the standard deviation. Percent cleavage is defined as (cleaved/(cleaved + uncleaved)) × 100. Percent Dna2 bound is defined as (Dna2 bound/(Dna2 bound + FEN1 bound + unbound substrate)) × 100. Percent FEN1 bound is defined as (FEN1 bound/(FEN1 bound + Dna2 bound + unbound substrate)) × 100. Substrates are depicted above gels with the RNA labeled in gray and DNA in black. The asterisk indicates the site of the 3′-32P radiolabel.