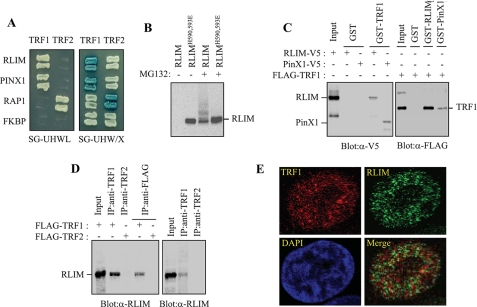
FIGURE 1.
Physical interaction between TRF1 and LRIM. A, analysis of the physical interaction between TRF1 and RLIM using the yeast two-hybrid assay. PinX1, RAP1, and unrelated FKBP52 were used as the TRF1-binding, TRF2-binding, and negative control, respectively. The growth on the SG-HWUL plate and the blue signal on the SG-HWU/X plate indicate activation of the reporter genes, LacZ and LEU2, respectively. S, synthetic; G, galactose; H, histidine (-); W, tryptophan (-); U, uracil, (-); L, leucine (-); X, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). B, H1299 cells were transfected with V5-RLIM or V5-RLIMH590,593E and either untreated or treated with 10 μm MG132 for 4 h. Cell lysates were analyzed by immunoblotting with anti-V5 antibody probe. C, interaction between TRF1 and RLIM in vitro. Left, GST and GST-TRF1 were immobilized on glutathione-Sepharose and incubated with exogenously expressed RLIM-V5 and PinX1-V5. After washing and SDS-PAGE, bound RLIM and PinX1 were detected by immunoblotting with anti-V5 antibody. Right, GST, GST-RLIM, and GST-PinX1 were incubated with exogenously expressed FLAG-TRF1 followed by immunoblotting with anti-FLAG antibody. D, coimmunoprecipitation of TRF1 and RLIM. Left, H1299 cells were transfected with FLAG-TRF1 or FLAG-TRF2 and then subjected to immunoprecipitation as indicated followed by immunoblotting with anti-RLIM antibody. Right, H1299 cells were subjected to immunoprecipitation with either anti-TRF1 or -TRF2 antibodies followed by immunoblotting with anti-RLIM antibody. E, H1299 cells were treated with 10 μm MG132 for 4 h and analyzed by indirect immunofluorescence. Paraformaldehyde-fixed cells were stained with anti-TRF1 (red) and anti-RLIM antibodies (green). DNA was stained with 4,6-diamidino-2-phenylindole (blue).