Abstract
The membrane-embedded rotor in the F0 sector of proton-translocating ATP synthases is formed from hairpin-like c-subunits that are protonated and deprotonated during energization of ATP synthesis. This study focuses on two c-subunit motifs that are unique to synthases of extremely alkaliphilic Bacillus species. One motif is the AXAXAXA sequence found in the N-terminal helix-1 instead of the GXGXGXG of non-alkaliphiles. Quadruple A→G chromosomal mutants of alkaliphilic Bacillus pseudofirmus OF4 retain 50% of the wild-type hydrolytic activity (ATPase) but <18% of the ATP synthase capacity at high pH. Consistent with a structural impact of the four alanine replacements, the mutant ATPase activity showed enhanced inhibition by dicyclohexylcarbodiimide, which blocks the helix-2 carboxylate. Single, double, or triple A→G mutants exhibited more modest defects, as monitored by malate growth. The key carboxylate is in the second motif, which is P51XXE54XXP in extreme alkaliphiles instead of the (A/G)XX(E/D)XXP found elsewhere. Mutation of Pro51 to alanine had been shown to severely reduce malate growth and ATP synthesis at high pH. Here, two Pro51 to glycine mutants of different severities retained ATP synthase capacity but exhibited growth deficits and proton leakiness. A Glu54 to Asp54 change increased proton leakiness and reduced malate growth 79-90%. The Pro51 and the Glu54 mutants were both more dicyclohexylcarbodiimide-sensitive than wild type. The results highlight the requirement for c-subunit adaptations to achieve alkaliphile ATP synthesis with minimal cytoplasmic proton loss and suggest partial suppression of some mutations by changes outside the atp operon.
Proton-translocating ATP synthases transport protons down their electrochemical gradient through the membrane-embedded F0 sector of the synthase. This facilitates use of the proton-motive force produced by the respiratory chain to drive ATP synthesis by a rotary mechanism during OXPHOS3 in both prokaryotes and eukaryotes (1-4). The critical proton-binding carboxylate residues on the F0 rotor are in the C-terminal helix-2 of the hairpin-like c-subunit. This subunit forms a ring that is composed of 10-15 c-subunit monomers depending on the organism (5-7). Helix-1 packs on the inside of the c-ring and helix-2 on the outside. Small residues in helix-1 are suggested to contribute to the tight packing seen in the c-ring structure (5), including a highly conserved GXGXGXG sequence (6, 8). On the outside of the ring, a single a-subunit binds to the ring in bacteria, making sequential contact with pairs of c-ring subunits as rotation of the ring proceeds (9, 10). Protons are taken up from the outside using a pathway involving a-subunit residues. These residues are proposed to form an aqueous channel between the external surface of the bacterial membrane and the entry point of protons on to the c-ring within the membrane (11, 12). The path of protons that return from the c-ring and complete their downhill movement into the cytoplasm is less-well defined and may be through a distinct aqueous channel comprised of residues from the a-subunit (13) or via a pathway that involves the c-subunit (14). Interaction of the essential a-subunit arginine with the carboxylate of the c-subunits is a key event in the protonation/deprotonation reactions at the a/c interface during rotation of the c-ring (15, 16). The functional properties of the c-subunit carboxylate on helix-2 are also affected by residues in helix-1 that are opposite the carboxylate. For example, mutations in Ala24 and Ile28 in the Escherichia coli c-subunit helix-1 make the ATP synthase resistant to dicyclohexylcarbodiimide (DCCD), a reagent that specifically reacts with the carboxylate (Asp61 in E. coli) resulting in inactivation of the enzyme (17, 18).
OXPHOS supported by the proton-coupled ATP synthase of alkaliphilic Bacillus species at pH 10.5 presents a set of challenges with respect to capture of protons, successful proton translocation to the cytoplasm, and proton retention in the cytoplasm. B. pseudofirmus OF4 maintains a cytoplasmic pH of 8.2 during growth in rigorously controlled continuous culture at pH 10.5 in medium containing malate, a non-fermentable substrate (19, 20). This successful pH homeostasis is critical to growth at highly alkaline pH, so these organisms require mechanisms to minimize proton leakiness or membrane potential decreases that result in proton loss (21). It has been suggested that, without any specific adaptations of the OXPHOS machinery itself, the challenges of proton capture in support of OXPHOS and pH homeostasis can be overcome by global features that foster proton movement along the outer surface of the membrane to the ATP synthase and to the antiporters that support pH homeostasis (22). However, we have shown that the pKa of the c-subunit carboxylate (Glu54) of B. pseudofirmus OF4 is higher than those reported for several other bacterial c-subunit carboxylates, a possible adaptation for proton acquisition and retention during rotation of the c-ring (23).
Another indication that specific adaptations of the proton-coupled ATP synthase are involved in OXPHOS by extreme alkaliphiles is the presence of a group of alkaliphile-specific residues and motifs in functionally important regions of the a- and c-subunits of the synthases. The sequences of these residues and motifs differ significantly from the consensus sequences in non-alkaliphilic Bacillus species and the well studied E. coli ATP synthase (24, 25). In an initial mutagenesis study, we changed six of the most striking sequence features of the a- and c-subunits in the chromosome of genetically tractable B. pseudofirmus OF4 to the sequences found in non-alkaliphilic Bacillus megaterium (26). B. megaterium was chosen because its a- and c-subunit sequences have the closest overall sequence similarity to those of the B. pseudofirmus OF4 subunits among non-extremophilic Bacillus species. The results supported the conclusion that the ability of alkaliphiles to capture and productively translocate protons through the ATP synthase depends upon the alkaliphile-specific motifs or residues.
One of the features flagged by the initial mutagenesis study as crucial for ATP synthesis at high pH was the Pro51 residue found in a C-terminal helix-2 in extreme alkaliphiles but not in modest alkaliphiles such as Bacillus sp. TA2.A1. Arechaga and Jones designated as an “alkaliphile motif” the P51XXEXXP57 sequence that begins with Pro51, includes the proton-binding Glu54 residue, and then ends with a second, conserved Pro57 (27) (see the alignment in Fig. 1). In the only mutagenesis study of an ATP synthase in its natural alkaliphile host, we changed Pro51 of B. pseudofirmus OF4 to alanine, the residue found in non-alkaliphilic Bacillus species (Fig. 1). The cP51A mutation resulted in a severe deficit in malate growth and ATP synthase capacity at pH 10.5 and a much smaller deficit at pH 7.5 (26). The goals of the current study were to expand the characterization of the motifs of the alkaliphile c-subunit, the heart of the rotary ATP synthase, to a broader panel of mutants. In the earlier study, a striking candidate for another c-subunit motif of extreme alkaliphiles, AXAXAXA, was not studied in detail, because, when it was changed to the B. megaterium GXGXGXG sequence, almost no ATP synthase was found in the alkaliphile membrane (26). Alkaliphile c-subunits typically have substitution of two or more alanines for the glycines of the conserved helix-1 GXGXGXG motif found in non-alkaliphiles. The version of the motif with the full four alanines is found only in the most extreme alkaliphiles, such as B. pseudofirmus OF4 (Fig. 1). In this study, we first constructed a double mutant of B. pseudofirmus OF4 in which both the AXAXAXA motif and the Pro51 of helix-2 were changed to the B. megaterium sequence, because it was possible that more favorable helix interactions in the double mutant would support a higher membrane level of synthase. If not, mutagenesis of single and multiple alanine residues of the AXAXAXA motif, rather than its entire replacement, would be conducted. More limited replacement of the motif might support higher synthase levels in the membrane so that we could evaluate whether the motif has an important role in alkaliphile OXPHOS. We also investigated the effects of additional mutations in the P51XXEXXP57 motif, including: (i) mutation of the unusual Pro51 to glycine, which is found in the moderate alkaliphile and thermophilic Bacillus sp. TA2.A1 as well as E. coli (Fig. 1); (ii) mutation of Glu54 to aspartate; and (iii) mutation of conserved Pro57 to glycine and alanine. The results establish a major role for the AXAXAXA motif in alkaliphile OXPHOS and greatly extend our appreciation of the adverse consequences of mutational changes in the PXXEXXP motif.
FIGURE 1.
Alignment of c-subunit of the F1F0-ATP synthase from Bacillus species and E. coli. The shaded organisms are the extreme alkaliphiles B. pseudofirmus OF4, B. halodurans C-125, and B. alcalophilus, the more moderately alkaliphilic B. clausii, and the moderate alkaliphile and thermophile Bacillus sp. TA2.A1. Residue numbers are given for the B. pseudofirmus OF4 (top) and the E. coli c-subunits (bottom). The AXAXAXA motif and PXXEXXP motif are shaded. An open box shows the residues that align with the Gly17 residue, which includes the Ala24 residue of E. coli. The NCBI gene accession numbers for the data shown are: B. pseudofirmus OF4 (accession number AAC08039); B. halodurans C-125 (accession number NP_244626); B. alcalophilus (accession number AAA22255); B. clausii KSM-K16 (accession number YP_177348); Bacillus sp. TA2.A1 (accession number AAQ10085); B. megaterium (accession number AAA82521); Geobacillus kaustophilus HTA426 (accession number YP_149216); B. subtilis subsp. subtilis str.168 (accession number NP_391567); and E. coli K12 substr. DH10B (accession number YP_001732558).
EXPERIMENTAL PROCEDURES
Bacterial Strains, Mutant Construction, and Growth Conditions—The wild-type and all mutant strains used in this study are listed in Table 1. The wild-type strain is a derivative of alkaliphilic B. pseudofirmus OF4 811M that has an EcoR1 site introduced into atpB in the same location as found in all the mutants used in the study (26). Mutations in the atpE gene that encodes the c-subunit were constructed in a cloned atpB-F fragment containing the EcoR1 site. The mutated fragments were then introduced into a mutant strain of B. pseudofirmus OF4 811M, designated ΔF0, that is deleted in the F0-encoding genes (ΔatpB-F). The two-step method used to regenerate the complete, mutant chromosomal atp operons in the ΔF0 strain is described in Ref. 26. Briefly, mutagenesis of atpB-F fragments was conducted using the GeneTailor™ Site-Directed Mutagenesis System (Invitrogen), in the low copy plasmid pMW118 (Nippon Gene, Toyama, Japan). The primers used for all the mutations made in this study are listed in supplemental Table S1. The mutant atpB-F constructs from pMW118 were digested with BamHI and KpnI and then ligated with pG+host4 (Appligene, Pleasanton, CA) previously digested with BamHI and KpnI. The ligation mixtures were used to transform E. coli XL-1 Blue MRF (Promega, Madison, WI), and selection was on Luria-Bertani broth containing 250 μg/ml erythromycin. Constructs in pG+host4 with the correct mutant sequences were introduced into the ΔF0 strain (ΔatpB-F). Homologous recombination introduced the mutations into the chromosomal atp locus. This recombination and concomitant loss of the pG+host4 plasmid were achieved by temperature selection methods described previously (28). The F0 segment of each mutant was entirely sequenced and verified to have only the desired mutation(s). These and other DNA sequence analyses were conducted at Genewiz, Inc. (South Plainfield, NJ). For each mutation, a panel of candidate recombinant colonies was tested for growth on malate at pH 7.5 and pH 10.5. For most of the mutants, all of the colonies of a panel showed very similar growth, and a representative colony was chosen for study. For a few mutants, the panel yielded two types of colonies (e.g. one type that exhibited more growth on malate at pH 7.5 than the other). For those mutants, a representative colony was chosen for each type, and the entire atp operon was sequenced (see “Results” and “Discussion” for further details). Growth of wild-type and mutant strains on semi-defined glucose or malate medium were determined as described previously (26). Briefly, cells were pre-grown on glucose at pH 7.5 and inoculated into comparable media with the carbon source and pH indicated in the text or figures. For glucose growth, the small amount of growth on yeast extract as the sole carbon source was subtracted from total growth assessed by A600 after 14 h at 30 °C. For malate growth, the small amount of growth exhibited by the ΔF0 strain on malate was subtracted from the growth of the other strains. All growth experiments were conducted in duplicate in at least two independent experiments.
TABLE 1.
B. pseudofirmus OF4 strains used in this study
|
Strain
|
Properties
|
Source
|
|---|---|---|
| Wild type | Derivative of B. pseudofirmus OF4 811 m into which an EcoRI site was introduced in atpB by silent mutation of nucleotides 484 and 487 | Ref. 27 |
| ΔFo | Deletion of atpB-F | Ref. 27 |
| Mutants in the c-subunit helix-1 motifa | ||
| cHx1 | Substitution of wild-type atpE residues 15-23 (VAGAIAVAI) with corresponding B. megaterium residues 15-23 (LGAGIGNGL) | Ref. 27 |
| cP51A | P51XXEXXP57→ A51XXEXXP57 | Ref. 27 |
| cHx1-cP51A | Double cHx1 and cP51A mutant | This study |
| cA16G | A16XAXAXA22→ G16XAXAXA22 | This study |
| cA18G | A16XAXAXA22→ A16XGXAXA22 | This study |
| cA20G | A16XAXAXA22→ A16XAXGXA22 | This study |
| cA22G | A16XAXAXA22→ A16XAXAXG22 | This study |
| cA1620G | A16XAXAXA22→ G16XAXGXA22 | This study |
| cA162022G | A16XAXAXA22→G16XAXGXG22 | This study |
| cA4G | A16XAXAXA22→G16XGXGXG22 | This study |
| cG17A | cG17A mutant | This study |
| Mutants in the c-subunit helix-2 motifa | ||
| cP51G | P51XXEXXP57→ G51XXEXXP57 | This study |
| cE54D | P51XXEXXP57→ P51XXDXXP57 | This study |
| cP57G | P51XXEXXP57→ P51XXEXXG57 | This study |
| cP57A | P51XXEXXP57→ P51XXEXXA57 | This study |
The residues in boldface and underlined are mutated residues.
Isolation of Everted Vesicles, Assays of Their Octylglucoside-stimulated ATPase, and Assessments of the Membrane Content of ATP Synthase Protein—Wild-type and mutant derivatives were grown at 30 °C in a semi-defined medium containing 0.1% yeast extract with mineral salts and buffered with 0.1 m Na2CO3/NaHCO3 at pH 10.5 or buffered with 0.1 m MOPS at pH 7.5 with 50 mm glucose or malate as the major carbon and energy source (26). Everted membrane vesicles were prepared from overnight cultures of the wild-type strain and mutant derivatives. The cells were washed with 50 ml of 50 mm Tricine-NaOH, pH 8.0/5 mm MgCl2, pelleted, and resuspended in 25 ml of French Press buffer containing 50 mm Tricine-NaOH, pH 8.0/5 mm MgCl2, a protease inhibitor tablet (Roche Applied Science), 1 mm phenylmethylsulfonyl fluoride, and a trace amount of DNase I (Roche Applied Science). The cells were broken in a French Press cell at 18,000 p.s.i. The broken cell suspensions were centrifuged at 15,000 × g for 10 min to precipitate unbroken cells and debris. The resulting supernatants were subjected to ultracentrifugation at 250,000 × g (Beckman Ti-60 rotor for 1.5 h at 4 °C) to pellet the everted membrane vesicles. The everted vesicles were suspended in 1 ml of 50 mm Tricine-NaOH, pH 8.0/5 mm MgCl2. These vesicles were used for determinations of protein content, octylglucoside (OG)-stimulated ATPase (representing the total uncoupled ATPase activity), and the levels of ATP synthase protein in the membrane. Protein content for this and other experiments was measured by the Lowry method using bovine serum albumin as the standard (29). OG-stimulated ATPase assays were conducted as described previously (26). For assessment of the ATP synthase content of the everted vesicle membranes, the protein complement of the vesicles was fractionated on 12% SDS-PAGE gels (30), and transferred electrophoretically to nitrocellulose. Western analyses were carried out by the chemiluminescence method according to the manufacturer's instructions (Pierce). The β-subunit of the F1F0 was detected using a monoclonal antibody against the E. coli β-subunit of F1F0 ATP synthase (Molecular Probes, Eugene, OR). For purposes of quantitation, image analysis was performed using ImageJ 1.40 software (rsbweb.nih.gov/ij/).
Assays of ATP Synthesis in ADP plus Pi-loaded Right-side-out Membrane Vesicles—Wild-type and mutant derivatives were grown to an A600 of 0.6 in the semi-defined medium containing 0.1 m Na2CO3/NaHCO3 at pH 10.5 with glucose as the energy source or with Na2CO3-buffered medium at pH 8.5 with malate as the energy source, as indicated in connection with specific experiments. Right-side out (RSO) ADP plus Pi-loaded membrane vesicles were prepared by methods described previously (31). After the wash step, the vesicles were suspended in 0.25 m sucrose, 20 mm potassium phosphate, pH 8.3, 5 mm sodium phosphate, pH 8.3, and 5 mm MgCl2. ATP synthesis reactions were carried out at room temperature as follows. The ADP plus Pi-loaded RSO membrane vesicles were diluted 1:20, to 500 μg of protein/ml, into either pH 7.5 buffer containing 25 mm sodium phosphate, 0.25 m sucrose, 5 mm MgCl2, and 200 mm K2SO4 or into pH 10.5 buffer containing 25 mm Na2CO3, 0.25 m sucrose, 5 mm MgCl2, and 200 mm K2SO4, and ATP synthesis was initiated by energizing the aerated vesicles with 10 mm ascorbate and 0.1 mm ascorbate-phenazine methosulfate. Samples (200 μl) of the reaction mixtures were removed at 10 s and transferred to fresh tubes containing 50 μl of ice-cold 30% perchloric acid. After neutralization, the ATP content was determined by the luciferin-luciferase method (32). For each experimental set, samples were also taken of unenergized vesicles for assessment of background ATP values. The amount of ATP synthesized was calculated from a standard curve.
Determination of the Cytoplasmic pH after a Shift in the External pH from 8.5 to 10.5—pH shift experiments were conducted as described previously (26, 28). Cells were grown overnight (50 ml) at pH 8.5 in either malate- or glucose-containing medium, as indicated in connection with specific experiments. An equal volume of fresh medium was added to the overnight culture, and the cells were grown for three more hours. Cells were harvested by centrifugation, washed with and then resuspended in 3 ml of pH 8.5 buffer containing 100 mm Na2CO3/NaHCO3, 1 mm MgSO4, 1 mm KH2PO4. These manipulations under de-energized conditions provide a period of equilibration of the cytoplasmic pH with the buffer at pH 8.5. The A600 of the suspensions was adjusted to 20, and then each cell sample was diluted 1:25 into pH 10.5 buffer containing 100 mm Na2CO3/NaHCO3 and 10 mm malate. The radioactive probe, [14C]methylamine (1.7 μm with 60 mCi/mmol), was added to the dilution buffer. For controls for nonspecific binding of the probe, a parallel set of samples also contained 10 μm gramicidin. After 10-min incubation, the diluted suspensions were filtered through GF/F filters (Whatman). The filters were then dried, and the radioactivity was assessed by liquid scintillation spectrometry. Values for the cytoplasmic pH were calculated from duplicate measurements of the outside pH and the transmembrane pH gradient (ΔpH). The ΔpH was determined as described earlier from the measured distribution of radiolabeled methylamine (33). At least three independent assays were conducted.
DCCD Inhibition of Unstimulated ATPase Activity—A malachite green phosphate detection method was used to measure the low ATPase activity of the ATP synthase in the absence of OG. The sensitivity of this method was sufficient to give substantial absorbance readings throughout a time course of 0-30 min under the conditions described below. The time course was found to be linear and yielded the same specific activity as was reported in an earlier study (26). In that study, the less sensitive LeBel et al. method (34) was used for measuring phosphate release, requiring longer ATPase reaction times and less dilution of the DCCD-treated sample into the reaction mixture. The combination of these two aspects of the LeBel-based DCCD inhibition assay meant that the DCCD was probably diluted insufficiently to prevent further DCCD inhibition during the enzyme assay. Thus, in the current study, the wild-type enzyme was inhibited 20-24% as compared with ∼50% determined previously. The assay procedure was based on that previously described (35-37). The malachite green reagent contained malachite green (0.081%, w/v), polyvinyl alcohol (2.32%, w/v), ammonium molybdate (5.72%, w/v, in 6 m HCl), and water, mixed in the ratio 2:1:1:2. The reagent was used after it turned to a golden-yellow color, from its initial dark brown color, as described. For assays of DCCD sensitivity, everted membrane vesicles were preincubated with 10 μl of methanol (no DCCD) or 10 μl of 10 mm DCCD (dissolved in methanol) to 100 μm final concentration in 1 ml at a concentration of 2.5 mg of protein/ml for 30 min at room temperature in 20 mm MOPS-NaOH, pH 7.0, 5 mm MgCl2. Aliquots of 200 μl of the preincubation mixture were then diluted 10-fold into pre-warmed assay buffer containing 50 mm Tricine-NaOH, pH 8.0, 5 mm MgCl2, 5 mm ATP. Reactions were carried out for 10 min at 37 °C; preliminary experiments showed that the reaction was linear for 30 min. The reactions were terminated by transferring 200 μl of the reaction mixture to a fresh tube containing 800 μl of malachite green reagent. After 1 min of color development, 100 μl of 34% sodium citrate was added to stop further color development, and the tubes were allowed to stand at room temperature for 15 min. The absorbance at 620 nm was measured and compared with a Pi standard curve.
RESULTS
Levels of ATP Synthase in a Mutant with the B. megaterium GXGXGXG Motif Replacing the Native Alkaliphile AXAXAXA Motif Are Not Significantly Enhanced by Also Introducing a cP51A Mutation—In the initial mutagenesis study of alkaliphile-specific c-subunit features, the AXAXAXA sequence in the N-terminal helix-1, which is V15AGAIAVAI23 in B. pseudofirmus OF4 when including two flanking residues, was replaced with the sequence found in the corresponding location of the B. megaterium c-subunit, L15GAGIGNGL23 (see Fig. 1). Very little ATP synthase was found in the membranes of that mutant. This made it impossible to assess whether there were functional consequences of replacing the alternating alanines of the alkaliphile helix-1 motif with glycines, as found in non-alkaliphile c-subunits (26). Here, we first tested the possibility that the B. megaterium sequence would be more compatible with normal levels of ATP synthase in the membrane if the Pro51 of the PXXEXXP sequence in the opposing helix-2 of the alkaliphile c-subunit was changed to alanine, the residue found in B. megaterium. As shown in Table 2, the double mutant grew well on glucose at pH 10.5, but could not grow on malate at either pH 7.5 or pH 10.5. The double mutant had a membrane β-subunit content (representing ATP synthase protein level) that was <10% of the wild-type level. Thus the low levels of the of the ATP synthase in the alkaliphile mutant with a replacement of the helix-1 AXAXAXA region with the complete B. megaterium sequence was not due to an effect of the PXXEXXP motif on the opposing helix. Subsequent mutant analyses (shown below) demonstrated that B. pseudofirmus OF4 mutants with glycine(s) replacing one, two, three, or four alanines of the alkaliphile motif all have significant membrane levels of ATP synthase. This suggested that other residues, i.e. residues surrounding the alternating alanines or glycines of helix-1, contributed to the deficit in ATP synthase in the membrane, because some of them are different in B. pseudofirmus OF4 than in B. megaterium. Those contributors could be the following: Leu15, which is Val15 in the alkaliphile; Ala17, which is Gly17 in the alkaliphile; Asn21, which is Val21 in the alkaliphile; and/or Leu23, which is Ile23 in the alkaliphile. To test the idea that these altered residues contributed to the loss of ATP synthase in the membrane with a total motif replacement, we constructed one additional mutant in which only the Gly17 of the alkaliphile motif was replaced by alanine, as is found in the same position in B. megaterium. That single change resulted in a 35% reduction in the level of ATP synthase in the membrane, relative to the wild-type level. This significant reduction was consistent with the notion that the “Xs” of the motif have an impact on membrane levels of the enzyme (Table 2).
TABLE 2.
A cHx1-cP51A double mutant is highly deficient in growth on malate and β-subunit content, and a single cG17A mutant alone exhibits significant defects
|
Growtha,b
|
β-Subunit content
(Western)a,c
|
||||
|---|---|---|---|---|---|
| Strain |
Glucose
|
Malate
|
|||
| pH 7.5 | pH 10.5 | pH 7.5 | pH 10.5 | ||
| % | |||||
| Wild-type | 100 | 100 | 100 | 100 | 100 |
| cHx1 | 105 ± 18 | 107 ± 10 | 0 | 0 | 4 ± 0.1 |
| cP51A | 62 ± 9 | 89 ± 13 | 75 ± 6 | 23 ± 10 | 73 ± 4 |
| cHx1-cP51A | 110 ± 3 | 101 ± 3 | 0 | 0 | 6 ± 0.4 |
| cG17A | 103 ± 7 | 76 ± 13 | 55 ± 18 | 60 ± 11 | 65 ± 12 |
% of wild-type.
Average of two independent experiments ± S.D.
Average of two vesicle preparations ± S.D.
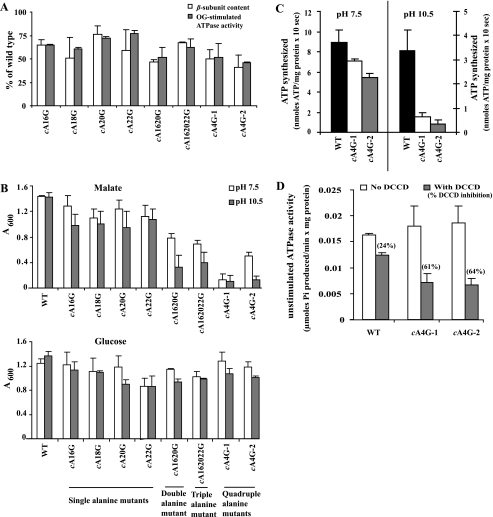
A Panel of AXAXAXA Mutations with Only A→G Changes Reveals a Crucial Role of the Motif in OXPHOS at High pH—A new mutant panel was constructed in the alkaliphile AXAX-AXA sequence that changed the alternating alanines without concomitant changes in residues surrounding the alanines. The panel included mutants with the following A→G changes (see Table 1): four single mutants, cA16G, cA18G, cA20G, and cA22G; a double mutant in the first and third alanines, cA1620G, which is the pattern found in the alkaliphilic B. clausii c subunit; a triple mutant in the first and the last two alanines, cA162022G; and the quadruple mutant, cA16182022G, which we designated cA4G. In preliminary studies of the quadruple mutant, we found two growth phenotypes even though sequence analyses showed that the F0 sequence of both types was identical and was the expected sequence. We chose a representative of each phenotype for inclusion in further studies, designated cA4G-1 and cA4G-2. The sequence of the entire atp operon in these strains showed that there were no mutations other than the four changes of alanine to glycine that we had introduced. All eight alkaliphile mutants in the new panel had over 40% of the ATP synthase levels found in wild type (Fig. 2A), in contrast to the alkaliphile mutants into which the B. megaterium sequence had been introduced (Table 2). The level of OG-stimulated ATPase activity, relative to that of wild-type, paralleled the ATP synthase content throughout the new mutant panel (Fig. 2A). Because OG-stimulated ATPase activity represents the total (uncoupled) hydrolytic activity, the specific hydrolytic activity was not significantly altered by any of the A→G mutations. All the mutants also grew well on glucose (≥63% of wild type) both at pH 7.5 and pH 10.5 (Fig. 2B, bottom). This was not the case for malate growth of all the mutants (Fig. 2B, top). All four mutant strains with only a single A→G substitution had a reproducible but small growth deficit relative to wild type on malate, with growth ≥76% and ≥66% of the wild-type at pH 7.5 and 10.5, respectively. The deficits in the double, triple, and quadruple mutants were much greater, especially at pH 10.5. The double and triple A→G replacement mutants exhibited malate growth that was 48-54% of wild type at pH 7.5 and only 23-27% at pH 10.5. The two quadruple mutant types, represented by strains cA4G-1 and cA4G-2, exhibited poor growth on malate. Both strains showed almost no growth on malate at pH 10.5 (7-9% of wild-type malate growth with no statistical difference between the two strains). At pH 7.5, malate growth of cA4G-2 was 35% of wild type, whereas malate growth of cA4G-1 was even worse, at only 9% of wild type.
FIGURE 2.
Functional characterization of helix-1 alanine mutants. A, β-subunit content and OG-stimulated ATPase activity of mutant strains. Strains were grown on glucose at pH 10.5. The values for the mutants are given as % of wild type, with the wild type set at 100%. Values are the average of determinations from at least two independent vesicle preparations, and the error bars show the ± S.D. B, growth of wild-type and mutant strains as a function of pH and carbon source. The values are the average of duplicate determinations from at least two independent growth experiments, and the error bars show the ±S.D. C, ATP synthesis by ADP plus Pi-loaded RSO membrane vesicles of wild type and the two types of quadruple A→G mutants. Vesicles were prepared from cells grown on glucose at pH 10.5. Assays were initiated by the addition of ascorbate-phenazine methosulfate as described under “Experimental Procedures.” The values are the average of duplicate assays from at least three independent vesicle preparations, and the error bars show the ±S.D. D, DCCD inhibition of unstimulated ATPase activity of wild-type and the two types of quadruple A→G mutants. Assays were conducted as described under “Experimental Procedures.” Values are the average of duplicate determinations from at least two independent vesicle preparations, and the error bars show the ±S.D. The numbers in parentheses above the gray columns indicate the % DCCD inhibition.
Further studies were conducted on the two quadruple alanine mutants cA4G-1 and cA4G-2 to assess the correlation of their phenotypes with their ATP synthesis activities (which we will call ATP synthase activities throughout the text) and with sensitivity of their ATPase activity to inhibition by DCCD. For in vitro assays of ATP synthesis, ADP plus Pi-loaded RSO vesicles with an intravesicular pH of 8.3 were prepared from wild-type and mutant cells. ATP synthesis was determined after energization of the vesicles with ascorbate-phenazine methosulfate at external pH values of 7.5 and 10.5. The intravesicular pH of 8.3 was comparable to the cytoplasmic pH of cells growing on malate at pH 10.5 (20). The shift of the vesicles loaded with buffer at pH 8.3 to either pH 7.5 or 10.5 imposed a pH gradient of 0.8 units, alkali in, at pH 7.5, whereas a 2.2-unit pH gradient, acid in, was imposed at pH 10.5. Consequently, the bulk proton electrochemical gradient was much higher at pH 7.5 in these experiments than at pH 10.5. The wild-type ATP synthase activities at pH 10.5 were 38% of those at pH 7.5 for vesicles prepared from glucose-grown cells (Fig. 2C) and 45% of those at pH 7.5 for vesicles prepared from malate-grown cells (see Fig. 3B). Consistent with the malate growth patterns at high pH, the ATP synthase activity of both A4G-1 and A4G-2 at pH 10.5 was very low (12-20% of wild-type) and not significantly different from each other, although that of A4G-1 always exhibited a little higher activity (Fig. 2C). Surprisingly, the ATP synthase activities of the two mutant strains at pH 7.5 were also not significantly different from each other, at 62-81% of wild type, despite the better malate growth of the cA4G-2 mutant at that pH (Fig. 2, B and C).
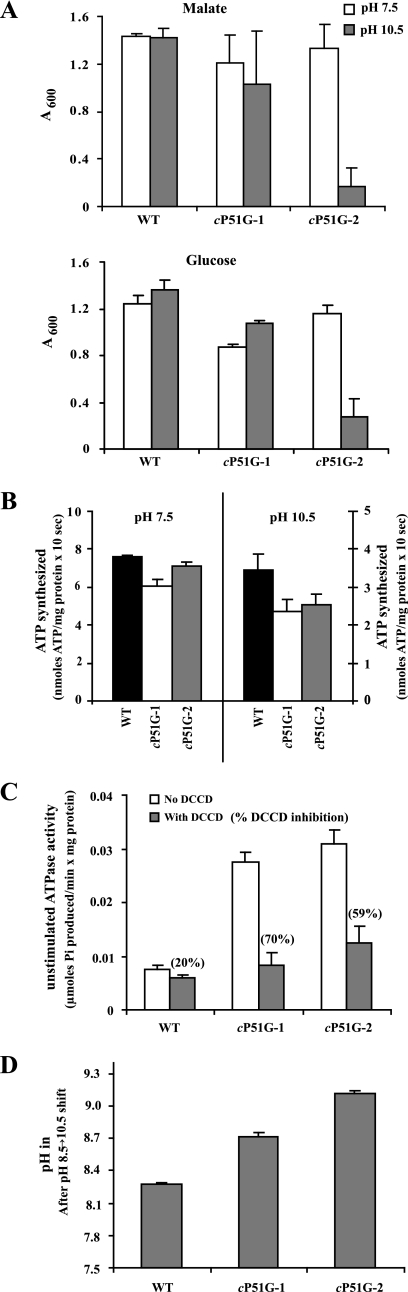
FIGURE 3.
Functional characterization of the cP51G mutants. A, growth of wild-type and the two types of cP51G mutants as a function of pH and carbon source. Assays were conducted as described earlier. The values are the average of duplicate determinations from at least two independent growth experiments, and the error bars show the ±S.D. B, ATP synthesis by ADP plus Pi-loaded RSO membrane vesicles of wild-type and the two types of cP51G mutants. Vesicles were prepared from cells grown on malate at pH 8.5. Assays were conducted as described under “Experimental Procedures.” The values are the average of duplicate assays from at least two independent vesicle preparations, and the error bars show the ±S.D. C, DCCD inhibition of unstimulated ATPase of the wild-type and the two types of cP51G mutants. Strains were grown on glucose at pH 7.5. Assays were conducted as described under “Experimental Procedures.” Values are the average of duplicate determinations from at least two independent vesicle preparations, and the error bars show the ±S.D. The numbers in parentheses above the gray columns indicate the % DCCD inhibition. D, determination of the cytoplasmic pH after a shift in the external pH from 8.5 to 10.5 of the wild type and the two types of cP51G mutants. Cells were grown on malate at pH 8.5. Assays were conducted as described as “Experimental Procedures.” Values are the average of duplicate determinations from at least three independent experiments, and the error bars show the ±S.D.
We then probed whether there were differences in the sensitivity of unstimulated ATPase activity to inhibition by DCCD between the two quadruple mutant strains and the wild type. We had shown earlier that the unstimulated hydrolytic activity of the purified alkaliphile ATP synthase reconstituted in proteoliposomes resulted in proton pumping that was abolished by DCCD pretreatment (and, additionally, that hydrolysis was stimulated by uncouplers) (38). Thus this unstimulated ATPase activity represents a coupled activity. DCCD inhibition of ATPase activity is the most widely used method to evaluate the reactivity of the crucial carboxylate with DCCD (see, e.g. Ref. 39). The extent of DCCD inhibition is expected to correlate with access of the carbodiimide reagent to the c-subunit carboxylate on helix-2, which in turn can be affected by changes in c-subunit structure. Alternatively, mutations in helix 1 could cause structural changes that affect the pKa of the carboxylate, resulting in altered reactivity with DCCD. The coupled ATPase activity of the wild-type B. pseudofirmus OF4 enzyme was inhibited 20-24% by DCCD (24% in the case of the cA4G mutants, Fig. 2D). The ATPase of both quadruple mutants with the AXAXAXA changed to GXGXGXG exhibited increased DCCD inhibition relative to that of the wild-type enzyme (61-64% inhibition for both mutant types, well over twice the sensitivity of wild type) (Fig. 2D). The results of the ATP synthase activity and DCCD inhibition of ATPase in the two quadruple mutants were consistent with an important role of the AXAX-AXA sequence for ATP synthase activity at pH 10.5 and with an impact of the quadruple mutation on accessibility of the c-ring carboxylate to DCCD and/or its reactivity with DCCD.
Mutants such as the two cA4G mutants that have major defects in alkaline growth need to be assessed for their relative proton leakiness, because a large pH gradient (more acid in the cytoplasm relative to the outside medium) is required for robust alkaliphile growth at high pH (21, 40). Our assay for this property was to determine the effect of an alkaline shift in the outside pH on the pH of the cytoplasm. Wild-type and quadruple mutant cells were grown on glucose at pH 8.5 (because malate growth of cA4G-1 was poor), washed, and equilibrated in pH 8.5 buffer and then shifted by dilution into malate and sodium-containing buffer at pH 10.5. The cytoplasmic pH values 10 min after the shift were 8.86 ± 0.04 for wild-type, 8.92 ± 0.01 for cA4G-1, and 8.68 ± 0.03 for cA4G-2. This indicated somewhat better pH homeostasis in cA4G-2 than in wild-type and slightly worse pH homeostasis in cA4G-1 than in wild-type, but neither mutant exhibited a large defect in pH homeostasis that would suggest leakiness. We note that the glucose-grown wild-type cells in these experiments showed a lower capacity for rigorous cytoplasmic pH homeostasis than is observed with malate-grown wild-type cells; malate-grown wild-type cells typically exhibit a cytoplasmic pH of 8.2-8.3 after such a shift (40, 41) (see “Discussion”).
Analyses of PXXEXXP Mutants in Alkaliphile-specific Pro51, the Carboxylate Glu54, and the Conserved Pro57 Show That Changes Result in Diverse Defects in the Alkaliphile Setting—The Pro51 that produces the P51XXEXXP57 motif (27) of extreme alkaliphiles is important for OXPHOS at high pH. This conclusion is based on the poor growth on malate and greatly reduced ATP synthase activity, at pH 10.5, of the P51A mutant characterized in our initial mutagenesis study; alanine is the residue found in the c-subunit 51-position in non-alkaliphilic Bacillus species (Fig. 1) (25, 26). The first new mutant of the PXXEXXP motif in this study again focused on Pro51 of B. pseudofirmus OF4 and changed Pro51 to glycine. Glycine is present at the position equivalent to the Pro51 of extreme alkaliphiles both in E. coli and in the more moderately alkaliphilic and thermophilic Bacillus TA2.A1 (42) (Fig. 1). Two types of phenotypes were observed among the group of cP51G mutants that were isolated. We chose strains that were representative of the two types and designated them cP51G-1 and cP51G-2. Sequence analyses showed that neither mutant had any mutations in the atp operon apart from the change introduced in Pro51. Both mutant strains had ≥73% of the ATP synthase level and ≥78% of the OG-stimulated ATPase activity of wild-type membranes (data not shown). cP51G-1 did not exhibit significant growth deficits relative to wild-type on either glucose or malate at either pH 7.5 or 10.5. By contrast, cP51G-2 also showed no significant growth defect on either carbon source at pH 7.5 but exhibited a large deficit in growth at pH 10.5 on both carbon sources, i.e. exhibited a non-alkaliphilic phenotype. Growth of cP51G-2 was only 12-20% of wild-type at high pH (Fig. 3A). The ATP synthase activities of the two mutant types did not correlate with their different growth phenotypes at pH 10.5, inasmuch as cP51G-1 and cP51G-2 both had ≥80% of the wild-type ATP synthase activity at pH 7.5 and had 69-76% of the wild-type activity at pH 10.5 (Fig. 3B). Membrane vesicles of both of the mutant strains exhibited a >3.5-fold increase in the unstimulated ATPase activity relative to wild type. Both mutants also showed a >2-fold increase in the % DCCD inhibition of that coupled ATPase activity, relative to wild type (Fig. 3C). A highly significant difference was observed between the two mutants and wild type in pH shift experiments that were conducted on malate-grown cells. Wild type exhibited a cytoplasmic pH of 8.3 10 min after a shift in external pH from 8.5 to 10.5, whereas the cytoplasmic pH values in the cP51G-1 and cP51G-2 mutants were, respectively, 8.7 and 9.1 (Fig. 3D). These assays also revealed the first difference between the two cP51G mutant strains that could relate to their different growth phenotypes, because the pH homeostasis defect of cP51G-2 was significantly greater than that of cP51G-1 (Fig. 3D).
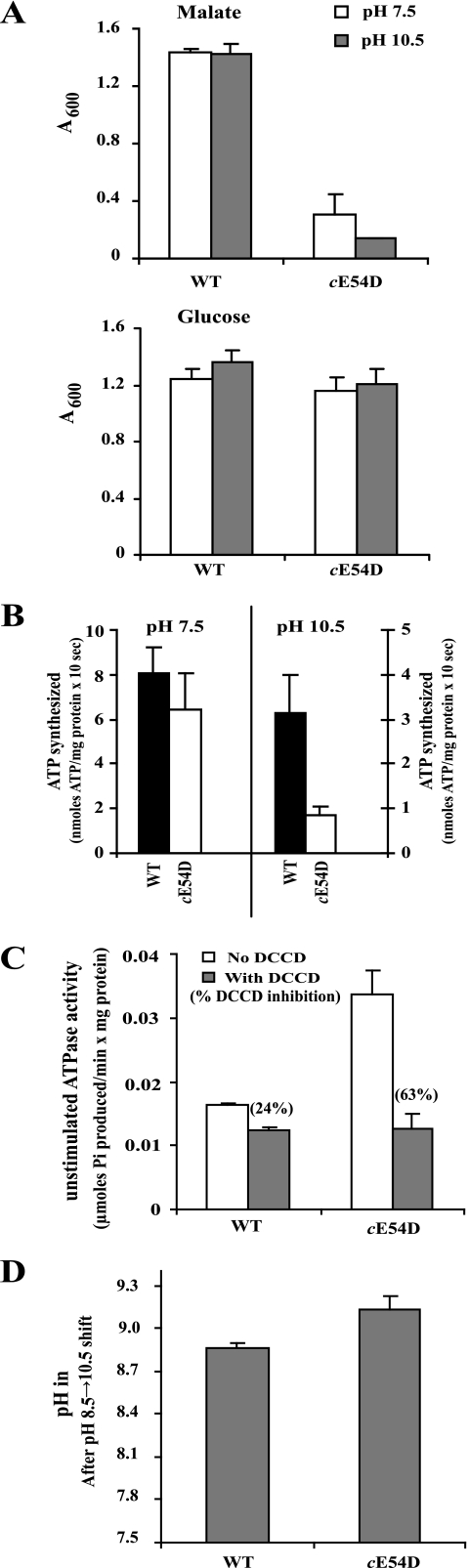
The second mutation made in the PXXEXXP motif of B. pseudofirmus OF4 was a change in the essential carboxylate, Glu54, to aspartate. Replacement by Miller et al. (43) of the corresponding carboxylate residue, Asp61, with a glutamate in E. coli was tolerated; the mutant retained ∼50% of the wild-type capacity for growth on succinate. The cE54D mutant of B. pseudofirmus OF4 grew as well as wild type on glucose at either pH 7.5 or 10.5 (Fig. 4A, bottom) and had wild-type levels of membrane ATP synthase β-subunit as well as OG-stimulated ATPase activity (data not shown). However, the mutant had a severe defect in malate medium, with growth yields of only 21 and 10% of wild-type at pH 7.5 and 10.5, respectively (Fig. 4A, top). The ATP synthase activity assayed in vitro was not significantly different from wild type at pH 7.5 despite the profound growth defect on malate at this pH (Fig. 4B). At pH 10.5, the ATP synthase activity of the cE54D mutant was 30% of the wild-type ATP synthase activity, thus correlating better with the very poor growth of the mutant at pH 10.5 (Fig. 4B). Assays of coupled ATPase activity showed that the activity in the cE54D mutant in the absence of added DCCD was more than twice that found in wild type, consistent with uncoupling. The inhibition by DCCD was ∼2.5 times that of wild type (Fig. 4C). Assessments of cytoplasmic pH homeostasis using the pH shift protocol had to be conducted on glucose-grown cells because of the growth phenotype of the mutant. The cytoplasmic pH 10 min after a pH 8.5→10.5 shift was 9.13 ± 0.03 for the cE54D mutant compared with 8.86 ± 0.04 for the wild-type, suggestive of proton leakiness in the mutant that was significantly greater than observed with the cA4G mutants under the same conditions (Fig. 4D).
FIGURE 4.
Functional characterization of cE54D mutant. A, growth of wild type and cE54D mutant as a function of pH and carbon source. The values are the average of duplicate determinations from at least two independent growth experiments, and the error bars show the ±S.D. B, ATP synthesis by ADP plus Pi-loaded RSO membrane vesicles of wild type and the cE54D mutant. Vesicles were prepared from cells grown with glucose at pH 10.5. Assays were conducted as described under “Experimental Procedures.” The values are the average of duplicate assays from at least three independent vesicle preparations, and the error bars show the ±S.D. C, DCCD inhibition of unstimulated ATPase activity of wild-type and the cE54D mutant. Everted vesicles were prepared from strains grown on glucose at pH 10.5. Values are the average of duplicate determinations from at least two independent vesicle preparations, and the error bars show the ±S.D. The numbers in parentheses above the gray columns indicate the % DCCD inhibition. D, determination of the cytoplasmic pH after a shift in the external pH from 8.5 to 10.5 of wild-type and the cE54D mutant. Cells were grown with glucose at pH 8.5. Values are the average of duplicate determinations from at least three independent experiments, with the wild-type data set serving as the control for these assays of cytoplasmic pH homeostasis as well as those of the cA4G mutants. The error bars show the ±S.D.
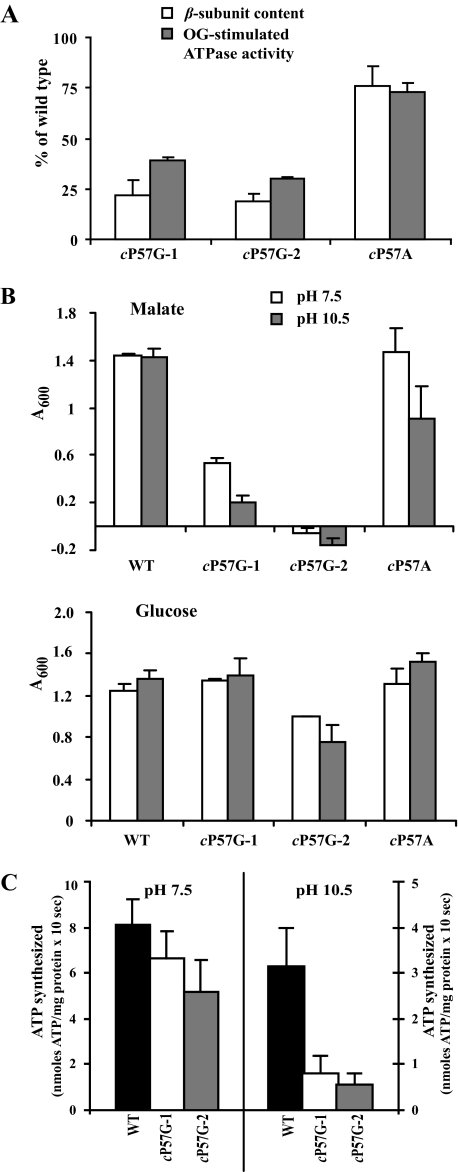
The second proline in the PXXEXXP motif is a conserved proline, in both alkaliphiles and neutrophiles, three residues from Glu54 on the C-terminal side. We constructed cP57A and cP57G mutations to test the importance of this proline in the B. pseudofirmus OF4 context. The cP57A mutant strain exhibited 76 and 73% of wild-type levels of enzyme and OG-stimulated ATPase activity, respectively (Fig. 5A). The growth of cP57A on either glucose or malate at either pH 7.5 or 10.5 was ≥64% of wild-type with only growth on malate at pH 10.5 showing any deficit (Fig. 5B). Two types of cP57G mutants were identified among the mutant isolates. Representative examples of the two types, designated cP57G-1 and cP57G-2, were shown by sequencing to have no mutations in the atp operon except for the cP57G change. Both cP57G-1 and cP57G-2 had greatly reduced membrane levels of the β-subunit of the ATP synthase and OG-stimulated ATPase activity (Fig. 5A); the Western assays did not show a statistically significant difference between the two mutants, but the ATPase activity of cP57G-2 was a little lower than that of cP57G-1. cP57G-1 grew as well as wild-type on glucose at pH 7.5 or 10.5, while showing reduced malate growth, especially at pH 10.5 (Fig. 5B). The phenotype of cP57G-2 was more extreme, in that it exhibited a growth deficit on glucose as well as total absence of growth on malate (when corrections were made for growth by the ΔF0 strain) (Fig. 5B). ATP synthase activity was assessed in the two cP57G mutants. Both mutants showed similar results for ATP synthesis. cP57G-1 and cP57G-2, respectively, exhibited 82 and 63% of wild-type activity at pH 7.5, and 27 and 20% of wild-type at pH 10.5 (Fig. 5C). The results indicated that the lower membrane levels of ATP synthase in the cP57G relative to wild-type led to deficits in ATP synthase and malate growth that were much larger at pH 10.5 than at pH 7.5. They were also larger in cP57G-2 than in cP57G-1.
FIGURE 5.
Functional characterization of the cP57G and cP57A mutants. A, β-subunit content and OG-stimulated ATPase activity of mutant strains. Strains were grown with glucose at pH 10.5. The values for the mutants are given as % of wild-type, with the wild-type set at 100%. Values are the average of determinations from at least two independent vesicle preparations, and the error bars show the ±S.D. B, growth of wild type, the two types of cP57G mutants, and the cP57A mutant as a function of pH and carbon source. The values are the average of duplicate determinations from at least two independent growth experiments, and the error bars show the ±S.D. C. ATP synthesis by ADP plus Pi-loaded RSO membrane vesicles of wild type and two types of cP57G mutants. Vesicles were prepared from cells grown on glucose at pH 10.5. Assays were conducted as described under “Experimental Procedures.” The values are the average of duplicate assays from at least three independent vesicle preparations, and the error bars show the ±S.D.
DISCUSSION
The results presented here strongly support the importance of alkaliphile-specific motifs for function of the ATP synthase c-subunit in OXPHOS at high pH. At pH 10.5, the bulk proton-motive force is much lower than at pH 7.5, because of the success with which secondary antiporters maintain a cytoplasmic pH that is over two units lower than the outside pH (21, 40, 44). We have long proposed that sequestered proton translocation between proton-pumping respiratory chain elements and the proton-consuming ATP synthase plays a critical role, perhaps enhanced by high membrane levels of cardiolipin and an unusually high content of amino acids with anionic side chains in external loops of many alkaliphile membrane proteins (40). Sequestration could be mediated by fast movement of protons along the membrane surface as has been demonstrated in other systems (45, 46) and by a “surface proton-motive” force of greater magnitude than the bulk proton-motive force (22, 47). It is further possible that OXPHOS in some settings involves direct dynamic interactions between respiratory chain complexes and the ATP synthase, as was recently shown for alkaliphile cytochrome oxidase and ATP synthase in a reconstituted system (48). This study extends initial evidence that alkaliphile-specific motifs that are predicted to be just outside the membrane surface (26) or within the membrane are adaptations of the ATP synthase machinery itself that are required for OXPHOS in the alkaliphile context. These findings negate the suggestion that properties of surface retention or rapid movement of protons can fully address the bioenergetic challenge of OXPHOS by alkaliphiles at high pH without adaptations in the synthase (22, 47). The results here demonstrate that the ATP synthase c-subunit is specially adapted to achieve inward proton translocation that energizes ATP synthesis while minimizing loss of protons from the rotor or cytoplasm to the alkaline bulk phase.
A major new finding is the demonstration that the alkaliphile-specific AXAXAXA sequence of N-terminal helix-1 of the c-subunit plays an indispensable role in ATP synthase at high pH and hence is a functional motif. Replacement of only two of the four alanines with glycines produced a significant deficit in non-fermentative growth on malate, i.e. growth was 25% the wild-type level (Fig. 2B). Replacement of all the alanines with glycines resulted in ≥90% deficit in malate growth and in ≥82% deficit in in vitro ATP synthase activity at high pH relative to wild-type. Moreover, the findings suggest that the specific residues that surround the alanines of the AXAXAXA motif have functional impact since replacement of the four alanines as well as the surrounding residues to those found in the B. megaterium GXGXGXG motif led to reduced membrane levels of the ATP synthase, but comparable reductions were not found in the cA4G strains that had no changes in those additional residues (Table 2).
It is interesting to note that in studies by others of helix-1 of the E. coli c-subunit, mutation of Ala24 (equivalent to Gly17 in the alkaliphile) to serine or of Ile28 (equivalent to Val21 in the alkaliphile) to valine or threonine increased resistance of the enzyme to DCCD (17, 18). This contrasts with the increased inhibition found here when mutations were introduced on a different face of the helix, in a different frame of the neutrophile GXGXGXG motif or alkaliphile AXAXAXA. The GXGXGXG motif that is present in non-alkaliphiles has been proposed by Vonck et al. (8) to function in accommodating the tight packing of the inner ring of N-terminal helices in the c-ring of Ilyobacter tartaricus. In the alkaliphile setting, changes of AXAXAXAto GXGXGXG seemed to have “opened up” the structure as indicated by the greater inhibition by DCCD (Fig. 2D), if greater accessibility of the carboxylate to DCCD accounts for this change. However, it is possible that the structure of the quadruple mutants is still “tight” and that the greater DCCD inhibition is caused by an effect of the mutation on the pKa of the carboxylate that increases its reactivity with DCCD. A change in the pKa could perturb the protonation/deprotonation properties of the enzyme and thus account for the poor synthesis. The cA4G strains show little or no defect in pH shift experiments, consistent with retention of a relatively tight structure at least as far as proton leakage is concerned. We hypothesize that the less severe phenotype of cA4G-2 relative to cA4G-1 results from suppression by a change outside the atp operon. For example, a modest increase in activity of a sodium-proton antiporter in cA4G-2 could account for its slightly better pH homeostasis after a shift to pH 10.5 and could also account for its better malate growth at pH 7.5, because a higher sodium motive force is needed to drive alkaliphile transport systems that are coupled to sodium (e.g. malate transport) at pH 7.5 than at pH 10.5 (21, 49).
To the extent that the AXAXAXA motif has effects on the packing of the c-subunits, it could be involved in supporting formation of a c-ring that has a higher stoichiometry of c-subunit monomers than that found in non-alkaliphilic Bacillus species. A high c-subunit stoichiometry could be a partial solution to the conundrum of robust alkaliphile OXPHOS at high pH despite the lower bulk proton-motive force (7, 50, 51). So far, however, elevated c-subunit stoichiometries have been reported but were not associated with AXAXAXA motifs. The tridecameric c-ring of the moderate alkaliphile and thermophile Bacillus sp. TA2.A1, which has a higher stoichiometry than that observed in several non-alkaliphilic bacterial c-rings, has an unusual GXSXGXS sequence in this position (50). The role of that N-terminal helix sequence in the c-subunit stoichiometry or in synthase function at alkaline pH and/or elevated temperature has not yet been reported for Bacillus TA2.A1. In two cyanobacteria, Synechocystis PCC6803-14 and Spirulina (Arthrospira) platensis, that have c-rotors with even higher stoichiometries, c14 and c15 rings, respectively, the c-subunit helix-1 motif has a GXGXGXG sequence (7, 51). Therefore an elevated c-subunit stoichiometry does not require the AXAX-AXA sequence found in extreme alkaliphiles, at least up to 15 monomers/ring. Thus, although direct evidence for or against an effect of the quadruple mutation on c-subunit stoichiometry should be sought, it seems more likely that the alkaliphile AXAXAXA motif functions in some other way, e.g. by influencing the pKa of the carboxylate on the opposing helix.
The second major finding of the current study is that mutational changes in both the Pro51 and the Glu54 of the alkaliphile PXXEXXP motif to residues that support functional ATP synthases in non-alkaliphiles are not compatible with function of the alkaliphile ATP synthase at high pH. The two types of cP51G mutants showed a unique pattern among alkaliphile ATP synthase mutants studied to date. The ATP synthase capacity of the two strains, in assays of ATP synthesis in ADP plus Pi-loaded vesicles, was not severely compromised in either cP51G-1 or cP51G-2 (Fig. 3B). One of the mutant types, cP51G-1, also lacked major deficits in growth on both malate and glucose relative to wild-type, although malate growth, especially at pH 10.5, was much more variable than typically observed. By contrast, the growth phenotype of the other mutant type, c-P51G-2, was severe at pH 10.5 on both glucose and malate, where the growth yields were 20 and 12%, respectively, of wild-type yields (Fig. 3A). The ATPase activity in the absence of OG was greatly increased in both cP51G mutant strains relative to wild type (Fig. 3C), suggesting that the ATPase was to some extent uncoupled in both mutants. This was consistent with the proton leakiness evidenced by the large alkalinization (to 8.7 and 9.1) of the cytoplasm of both cP51G mutant strains after a shift in the external pH from 8.5 to 10.5; the wild-type cytoplasmic pH was 8.3 after the shift (Fig. 3D). A slightly larger unstimulated ATPase activity and cytoplasmic alkalinization were observed with cP51G-2 than with cP51G-1, but the differences seem too small to account for the much more severe phenotype of cP51G-2, especially at pH 10.5 (Fig. 3A). The effects of the mutation that were observed in the cP51G-2 strain may be suppressed by a change outside of the atp operon that ameliorates those effects in cP51G-1. As with the cA4G mutants, it will be of interest to identify the mechanism of such suppression, for which a change in the antiporter activity profile would again be a candidate.
Mutation of c-subunit Glu54 to aspartate yielded a single mutant type whose growth on glucose was comparable to wild type at both pH 7.5 and 10.5, but showed a large defect in malate growth at both pH values, with a greater deficit at pH 10.5 than at pH 7.5 (Fig. 4A). The cE54D mutant of the alkaliphile showed a larger deficit in non-fermentative growth (10-21% of wild type) than the 50% reported for a cD61E mutant in the carboxylate of the E. coli c-subunit (43). The ATP synthase activity of the alkaliphile cE54D mutant was also greatly reduced at pH 10.5 relative to wild type (Fig. 4B). The cE54D mutant had a large increase in unstimulated ATPase activity (that was accompanied by increased DCCD inhibition) (Fig. 4C) and also showed significant alkalinization of the cytoplasm in a pH shift experiment (Fig. 4D) relative to wild type. Such proton leakiness would be highly detrimental to non-fermentative growth, especially at pH 10.5. It is possible that the change in the pKa of the carboxylate also contributes to the deficit in ATP synthesis and non-fermentative growth, in view of the high pKa measured for the wild-type carboxylate (23). In sum, the findings with the cP51G and cE54D mutant strains underscore the role of alkaliphile-specific features of the PXXEXXP motif in protecting the native host from cytoplasmic proton loss. As with the helix-1 AXAXAXA mutations, more structural-functional information on the alkaliphile c-ring will be required before we fully understand all the current observations on the PXXEXXP motif.
The other new mutant in this study was in the conserved Pro57 of the PXXEXXP motif. Mutation of this residue to alanine did not produce a severe growth phenotype, whereas mutation to glycine produced two mutant types with respect to the severity of their growth phenotypes. Both types, represented by cP57G-1 and cP57G-2, exhibited major defects in the ATP synthase content and hence also had deficits in the OG-stimulated ATPase activity and in malate growth. Although the levels of these parameters were only slightly lower in cP57G-2 than in cP57G-1 (Fig. 5, A and C), the former strain did not grow on malate at all and also showed a modest growth deficit on glucose, whereas cP57G-1 showed no growth deficit on glucose and a smaller one on malate (Fig. 5B). Because no differences between the two mutants were found within the atp operon, suppression of the more severe phenotype of cP51G-1 may involve a change elsewhere. Clearly, though, the conserved Pro57 can be changed to either alanine or glycine without complete loss of ATP synthase activity.
In this study, we have used both glucose-grown and malate-grown cells for pH shift experiments in which we assessed effects of mutations on cytoplasmic pH homeostasis as an indicator of proton leakiness in ATP synthase F0 mutants. Glucose-grown cells had to be used for mutants that could not grow well on malate even at pH 8.5, i.e. the cA4G mutants and the cE54D mutant. We note that 10 min after a shift of glucose-grown cells of wild-type from the pH 8.5 equilibration buffer to the pH 10.5 buffer, the cytoplasmic pH was pH 8.86, whereas that of malate-grown cells was pH 8.27. Glucose-grown cells have lower levels of respiratory chain components and ATP synthase than malate-grown cells.4 This probably accounts for the reduced capacity for pH homeostasis following a sudden upward pH shift. It is also likely that proton capture during OXPHOS plays a role in pH homeostasis under alkaline conditions (21). Up-regulation of ATP synthase genes under alkali stress has been observed in E. coli (52), Bacillus subtilis (53), and Desulfovibrio vulgaris (54).
Supplementary Material
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s).
This work was supported, in whole or in part, by National Institutes of Health Grant GM28454 (to T. A. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: OXPHOS, oxidative phosphorylation; DCCD, dicyclohexylcarbodiimide; MOPS, 4-morpholinepropanesulfonic acid; OG, octylglucoside; RSO, right-side out; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
J. Liu, M. Fujisawa, D. B. Hicks, and T. A. Krulwich, unpublished data.
References
- 1.Mitchell, P. (1961) Nature 191 144-148 [DOI] [PubMed] [Google Scholar]
- 2.Boyer, P. D. (1997) Annu. Rev. Biochem. 66 717-749 [DOI] [PubMed] [Google Scholar]
- 3.Stock, D., Gibbons, C., Arechaga, I., Leslie, A. G., and Walker, J. E. (2000) Curr. Opin. Struct. Biol. 10 672-679 [DOI] [PubMed] [Google Scholar]
- 4.Yoshida, M., Muneyuki, E., and Hisabori, T. (2001) Nat. Rev. Mol. Cell Biol. 2 669-677 [DOI] [PubMed] [Google Scholar]
- 5.Stock, D., Leslie, A. G., and Walker, J. E. (1999) Science 286 1700-1705 [DOI] [PubMed] [Google Scholar]
- 6.Meier, T., Polzer, P., Diederichs, K., Welte, W., and Dimroth, P. (2005) Science 308 659-662 [DOI] [PubMed] [Google Scholar]
- 7.Pogoryelov, D., Yu, J., Meier, T., Vonck, J., Dimroth, P., and Muller, D. J. (2005) EMBO Rep. 6 1040-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vonck, J., von Nidda, T. K., Meier, T., Matthey, U., Mills, D. J., Kuhlbrandt, W., and Dimroth, P. (2002) J. Mol. Biol. 321 307-316 [DOI] [PubMed] [Google Scholar]
- 9.Mellwig, C., and Bottcher, B. (2003) J. Biol. Chem. 278 18544-18549 [DOI] [PubMed] [Google Scholar]
- 10.Rubinstein, J. L., Walker, J. E., and Henderson, R. (2003) EMBO J. 22 6182-6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angevine, C. M., and Fillingame, R. H. (2003) J. Biol. Chem. 278 6066-6074 [DOI] [PubMed] [Google Scholar]
- 12.Angevine, C. M., Herold, K. A., and Fillingame, R. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13179-13183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fillingame, R. H., Angevine, C. M., and Dmitriev, O. Y. (2003) FEBS Lett. 555 29-34 [DOI] [PubMed] [Google Scholar]
- 14.von Ballmoos, C., Cook, G. M., and Dimroth, P. (2008) Annu. Rev. Biophys. 37 43-64 [DOI] [PubMed] [Google Scholar]
- 15.Hatch, L. P., Cox, G. B., and Howitt, S. M. (1995) J. Biol. Chem. 270 29407-29412 [DOI] [PubMed] [Google Scholar]
- 16.Valiyaveetil, F. I., and Fillingame, R. H. (1997) J. Biol. Chem. 272 32635-32641 [DOI] [PubMed] [Google Scholar]
- 17.Hoppe, J., Schairer, H. U., and Sebald, W. (1980) Eur. J. Biochem. 112 17-24 [DOI] [PubMed] [Google Scholar]
- 18.Fillingame, R. H., Oldenburg, M., and Fraga, D. (1991) J. Biol. Chem. 266 20934-20939 [PubMed] [Google Scholar]
- 19.Guffanti, A. A., and Hicks, D. B. (1991) J. Gen. Microbiol. 137 2375-2379 [DOI] [PubMed] [Google Scholar]
- 20.Sturr, M. G., Guffanti, A. A., and Krulwich, T. A. (1994) J. Bacteriol. 176 3111-3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krulwich, T. A., Hicks, D. B., Swartz, T .H., and Ito, M. (2007) in Physiology and Biochemistry of Extremophiles (Gerday, C., and Glansdorff, N., eds) pp. 311-329, ASM Press, Washington, D. C.
- 22.Mulkidjanian, A. Y., Cherepanov, D. A., Heberle, J., and Junge, W. (2005) Biochemistry (Mosc) 70 251-256 [DOI] [PubMed] [Google Scholar]
- 23.Rivera-Torres, I. O., Krueger-Koplin, R. D., Hicks, D. B., Cahill, S. M., Krulwich, T. A., and Girvin, M. E. (2004) FEBS Lett. 575 131-135 [DOI] [PubMed] [Google Scholar]
- 24.Ivey, D. M., and Krulwich, T. A. (1991) Mol. Gen. Genet. 229 292-300 [DOI] [PubMed] [Google Scholar]
- 25.Ivey, D. M., and Krulwich, T. A. (1992) Res. Microbiol. 143 467-470 [DOI] [PubMed] [Google Scholar]
- 26.Wang, Z., Hicks, D. B., Guffanti, A. A., Baldwin, K., and Krulwich, T. A. (2004) J. Biol. Chem. 279 26546-26554 [DOI] [PubMed] [Google Scholar]
- 27.Arechaga, I., and Jones, P. C. (2001) FEBS Lett. 494 1-5 [DOI] [PubMed] [Google Scholar]
- 28.Ito, M., Guffanti, A. A., Zemsky, J., Ivey, D. M., and Krulwich, T. A. (1997) J. Bacteriol. 179 3851-3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265-275 [PubMed] [Google Scholar]
- 30.Schagger, H., and von Jagow, G. (1987) Anal. Biochem. 166 368-379 [DOI] [PubMed] [Google Scholar]
- 31.Guffanti, A. A., and Krulwich, T. A. (1994) J. Biol. Chem. 269 21576-21582 [PubMed] [Google Scholar]
- 32.Stanley, P. E., and Williams, S. G. (1969) Anal. Biochem. 29 381-392 [DOI] [PubMed] [Google Scholar]
- 33.Krulwich, T. A., Federbush, J. G., and Guffanti, A. A. (1985) J. Biol. Chem. 260 4055-4058 [PubMed] [Google Scholar]
- 34.LeBel, D., Poirier, G. G., and Beaudoin, A. R. (1978) Anal. Biochem. 85 86-89 [DOI] [PubMed] [Google Scholar]
- 35.Rowlands, M. G., Newbatt, Y. M., Prodromou, C., Pearl, L. H., Workman, P., and Aherne, W. (2004) Anal. Biochem. 327 176-183 [DOI] [PubMed] [Google Scholar]
- 36.Chan, K. M., Delfert, D., and Junger, K. D. (1986) Anal. Biochem. 157 375-380 [DOI] [PubMed] [Google Scholar]
- 37.Maehama, T., Taylor, G. S., Slama, J. T., and Dixon, J. E. (2000) Anal. Biochem. 279 248-250 [DOI] [PubMed] [Google Scholar]
- 38.Hicks, D. B., and Krulwich, T. A. (1990) J. Biol. Chem. 265 20547-20554 [PubMed] [Google Scholar]
- 39.Valiyaveetil, F., Hermolin, J., and Fillingame, R. H. (2002) Biochim. Biophys. Acta 1553 296-301 [DOI] [PubMed] [Google Scholar]
- 40.Krulwich, T. A. (1995) Mol. Microbiol. 15 403-410 [DOI] [PubMed] [Google Scholar]
- 41.Krulwich, T. A., Ito, M., Gilmour, R., and Guffanti, A. A. (1997) Extremophiles 1 163-169 [DOI] [PubMed] [Google Scholar]
- 42.Olsson, K., Keis, S., Morgan, H. W., Dimroth, P., and Cook, G. M. (2003) J. Bacteriol. 185 461-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, M. J., Oldenburg, M., and Fillingame, R. H. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 4900-4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Padan, E., Bibi, E., Ito, M., and Krulwich, T. A. (2005) Biochim. Biophys. Acta 1717 67-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heberle, J., Riesle, J., Thiedemann, G., Oesterhelt, D., and Dencher, N. A. (1994) Nature 370 379-382 [DOI] [PubMed] [Google Scholar]
- 46.Branden, M., Sanden, T., Brzezinski, P., and Widengren, J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19766-19770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulkidjanian, A. Y., Heberle, J., and Cherepanov, D. A. (2006) Biochim. Biophys. Acta 1757 913-930 [DOI] [PubMed] [Google Scholar]
- 48.Liu, X., Gong, X., Hicks, D. B., Krulwich, T. A., Yu, L., and Yu, C. A. (2007) Biochemistry 46 306-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilmour, R., Messner, P., Guffanti, A. A., Kent, R., Scheberl, A., Kendrick, N., and Krulwich, T. A. (2000) J. Bacteriol. 182 5969-5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meier, T., Morgner, N., Matthies, D., Pogoryelov, D., Keis, S., Cook, G. M., Dimroth, P., and Brutschy, B. (2007) Mol. Microbiol. 65 1181-1192 [DOI] [PubMed] [Google Scholar]
- 51.Pogoryelov, D., Reichen, C., Klyszejko, A. L., Brunisholz, R., Muller, D. J., Dimroth, P., and Meier, T. (2007) J. Bacteriol. 189 5895-5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer, L. M., Yohannes, E., Bondurant, S. S., Radmacher, M., and Slonczewski, J. L. (2005) J. Bacteriol. 187 304-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kosono, S., Asai, K., Sadaie, Y., and Kudo, T. (2004) FEMS Microbiol. Lett. 232 93-99 [DOI] [PubMed] [Google Scholar]
- 54.Stolyar, S., He, Q., Joachimiak, M. P., He, Z., Yang, Z. K., Borglin, S. E., Joyner, D. C., Huang, K., Alm, E., Hazen, T. C., Zhou, J., Wall, J. D., Arkin, A. P., and Stahl, D. A. (2007) J. Bacteriol. 189 8944-8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.