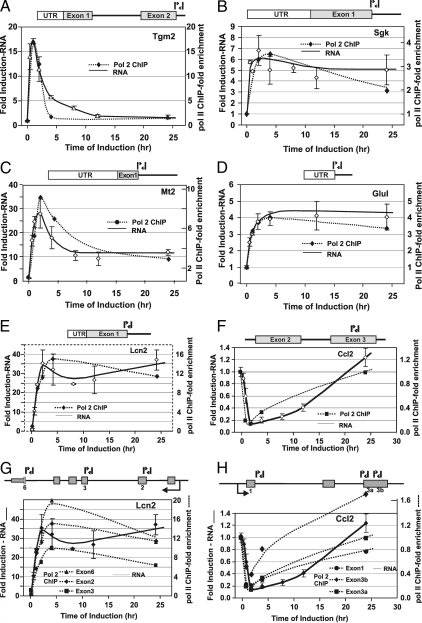
Figure 4.
Time-dependent occupancy of RNA pol II at GR-regulated genes in 3134. Relative pol II occupancy at selected genes was determined by ChIP analysis and compared with levels of transcription. Values are presented as a loading ratio relative to the amount determined before hormone treatment. A, Tgm2: pol II occupancy (-♦-) over a 24-h period of hormone treatment is compared with nascent RNA (—) levels. B, Sgk: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. C, Mt2: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. D, Glul: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. E, Lcn2: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. F, Ccl2: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. G, Lcn2: pol II binding (-♦-, -▴-, -▪-) over 24 h hormone treatment is monitored using multiple intron-exon primers across the transcribed locus and is compared with nascent RNA (—) levels. H, Ccl2: pol II binding (-♦-, -•-, -▪-) over 24 h hormone treatment is monitored using multiple intron-exon primers across the transcribed locus and is compared with nascent RNA (—-) levels. Error bars show the sd from the mean. ChIP q-PCR experiments represent at least two independent experiments (biological replicates) performed in duplicate (technical replicates). Primer locations are highlighted in the genomic structure diagrams for each locus.