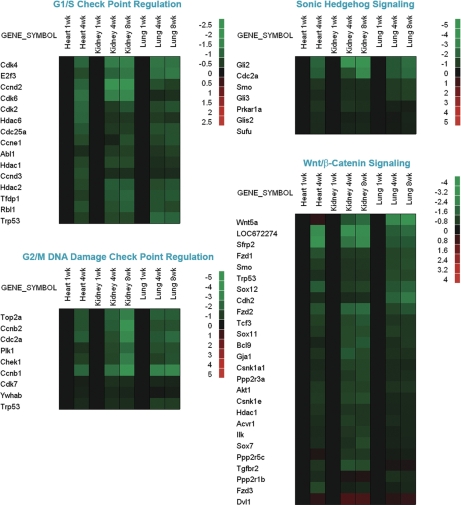
Figure 2.
Temporal changes in expression of genes participating in specific functional pathways. Microarray analysis was used to evaluate changes in gene expression in lung and kidney of 1-, 4-, and 8-wk-old mice and in heart of 1- and 4-wk-old mice. Ingenuity Pathways Analysis software was used to analyze changes in functional pathways as defined by that software package. Changes within a pathway were then visualized using heat maps constructed using JMP software version 7 (SAS Institute). To depict temporal changes in gene expression, the microarray values for each organ were normalized to the values for the same organ at 1 wk of age. Black rectangles represent no change in gene expression. Green rectangles represent down-regulated genes, and red rectangles represent up-regulated genes compared with 1-wk-old animals. The color intensity corresponds to the magnitude of the change from baseline, expressed as log2 (value at time point/value at 1 wk). Genes showing significant changes in expression with age (P < 0.05) in all three organs are depicted. The canonical pathways shown here are: G1/S check point regulation, G2/M DNA damage check point regulation, Sonic Hedgehog signaling, and Wnt/β-catenin signaling.