Abstract
The RAG1 and RAG2 proteins catalyze the site-specific DNA cleavage reactions in V(D)J recombination, the process that assembles antigen receptor genes from component gene segments during lymphocyte development. The first step towards the DNA cleavage reaction is the sequence-specific association of the RAG proteins with the conserved recombination signal sequence (RSS), which flanks each gene segment in the antigen receptor loci. Questions remain as to the contribution of each RAG protein to recognition of the RSS. For example, while RAG1 alone is capable of recognizing the conserved elements of the RSS, it is not clear if or how RAG2 may enhance sequence-specific associations with the RSS. To shed light on this issue, we examined the association of RAG1, with and without RAG2, to consensus RSS versus non-RSS substrates using fluorescence anisotropy and gel mobility shift assays. The results indicate that while RAG1 can recognize the RSS, the sequence-specific interaction at physiological conditions is masked by a high affinity non-sequence specific DNA-binding mode. Significantly, addition of RAG2 effectively suppressed the association of RAG1 with non-sequence specific DNA, resulting in a large differential in binding affinity for the RSS versus non-RSS sites. We conclude that this represents a major means by which RAG2 contributes to the initial recognition of the RSS, and that therefore association of RAG1 with RAG2 is required for effective interactions with the RSS in developing lymphocytes.
Keywords: protein-DNA interactions, V(D)J recombination, gel mobility shift assay, fluorescence anisotropy, recombination signal sequence
INTRODUCTION
RAG1 and RAG2 comprise the site-specific endonuclease that creates DNA double strand breaks in the initial steps of V(D)J recombination.1,2 Completion of the recombination process results in the combinatorial assembly of component gene segments to yield functional antigen receptor genes, an essential requirement for maturation of lymphocytes. The precision of the recombination events occurs through the site-specific recognition by the RAG proteins of the recombination signal sequence (RSS). Each gene segment in the antigen receptor loci that may be selected for recombination is flanked by an RSS. The RSS contains two conserved sequences, a heptamer and nonamer with consensus sequences of CACAGTG and ACAAAAACC, respectively. There are two types of RSS (12-RSS and 23-RSS) that are distinguished by the length of spacer sequence between the heptamer and nonamer. The RAG proteins associate with a 12-RSS and 23-RSS in a single complex (12/23 paired complex) as a prerequisite to completion of double-stranded cleavage between each RSS and its flanking gene segment.3–5 Thus, recombination between two gene segments is restricted to those flanked by RSS of differing spacer lengths, a phenomenon referred to as the 12/23 rule. Following site-specific DNA cleavage by the RAG proteins, the gene segments are joined by factors that function in the non-homologous end joining (NHEJ) DNA repair pathway to complete the V(D)J recombination reaction.6,7
RAG-mediated cleavage at each RSS occurs via two concerted catalytic steps.8 First, the RAG proteins nick DNA at the border between the RSS heptamer and the flanking gene segment. Second, the resulting 3′ hydroxyl group is activated to carry out a nucleophilic attack on the complementary strand of DNA, forming a DNA hairpin. Therefore, the products remaining after creation of each double strand break include the RSS with a 5′-phosphorylated blunt end and the gene segment terminated by a covalently sealed hairpin, termed signal and coding ends, respectively. Hairpin formation occurs only in the context of the 12/23 paired complex; however, nicking can occur at a single RSS.9–11
The assembly of the 12/23 paired complex appears to occur by a well-ordered series of macromolecular associations that is best represented by the capture model.11 In this model the RAG proteins appear to first bind and nick the 12-RSS. This is followed by recruitment of the 23-RSS, completion of nicking, and coupled hairpin formation at the two RSS to complete the cleavage of the double-stranded DNA. The capture model emphasizes the importance of the RAG proteins and 12-RSS interaction as this complex initiates V(D)J recombination. However, how the RAGs promote this site-specific interaction is not well understood. To understand the formation of the RAG:12-RSS complex, it is important to elucidate the contribution of each RAG protein to 12-RSS recognition.
Murine RAG1 and RAG2 are 1040 and 527 residues in length, respectively. Truncated forms of both proteins, referred to as core RAG1 (residues 384–1008) and core RAG2 (residues 1–387), are the minimal regions required to catalyze V(D)J recombination both in vitro and in vivo.2 RAG1 contains binding sites for both the heptamer and nonamer of the RSS, as determined by DNA footprinting12 and competition assays.13,14 Initial association with the RSS is believed to be through recognition of the nonamer element, which is driven largely through the N-terminal region (residues 384–470) of core RAG1.15,16 This region of core RAG1 is referred to as the nonamer binding region (NBR). Recognition of the RSS heptamer is mediated by the core RAG1 central domain (residues 528–760),17–19 with effective interaction most likely requiring distortion of the double-stranded B-form helix at the heptamer/flanking DNA.13,20–23
It is not yet resolved if RAG1 alone is sufficient to mediate sequence-specific interactions with the RSS, as previous studies of the RAG1:RSS interaction have produced contradictory conclusions. For example, it has been concluded that RAG1 forms mainly non-sequence specific interactions with DNA in the absence of RAG2.24 In contrast, an opposing view is that RAG1 alone could conceivably form the initial interactions with the RSS during V(D)J recombination.25 It has also been suggested that RAG2 enhances sequence-specific association of RAG1 with the RSS by inducing a conformational change in RAG1, although the nature of the conformational change and how it affects DNA binding is not known.
To gain insight into the relative roles of the RAG proteins in binding to the RSS, we investigated the binding affinity of RAGs to the consensus 12-RSS versus non-sequence specific (or non-specific) DNA substrates using both fluorescence anisotropy and electrophoretic mobility shift assays (EMSA). Our findings show that RAG1 alone exhibits sequence-specific DNA-binding character, but that this property is masked by a highly cooperative non-specific DNA binding interaction. Significantly, RAG2 inhibits the non-specific DNA binding mode of RAG1 to promote high affinity sequence-specific interactions with 12-RSS. We propose that this is a major mechanism by which the RAG2 component of the V(D)J recombinase helps to establish initial contact with the 12-RSS.
RESULTS
MCR1 binds with high affinity to both the consensus RSS and non-specific DNA
Core RAG1 was previously reported to bind with sequence specificity to the consensus 12-RSS as determined by various methods, including DNA footprinting12,13,22 and competition assays monitored by EMSA.13,14 Each method demonstrated that core RAG1 discriminated the nonamer sequence from non-specific DNA, with weak recognition of the double-stranded RSS heptamer sequence. However, the extent to which core RAG1 distinguishes consensus 12-RSS from non-specific DNA is in dispute in the literature.13,14,24,25 To clarify this issue, the relative binding energetics were determined for the interaction of MCR1 (core RAG1 fused to the C-terminal end of maltose binding protein) with a wild type (WT) 12-RSS substrate versus a non-specific substrate. The non-specific substrate used here, termed MHMN, is similar in both length and sequence to the non-specific DNA substrates used in previous studies.14,25
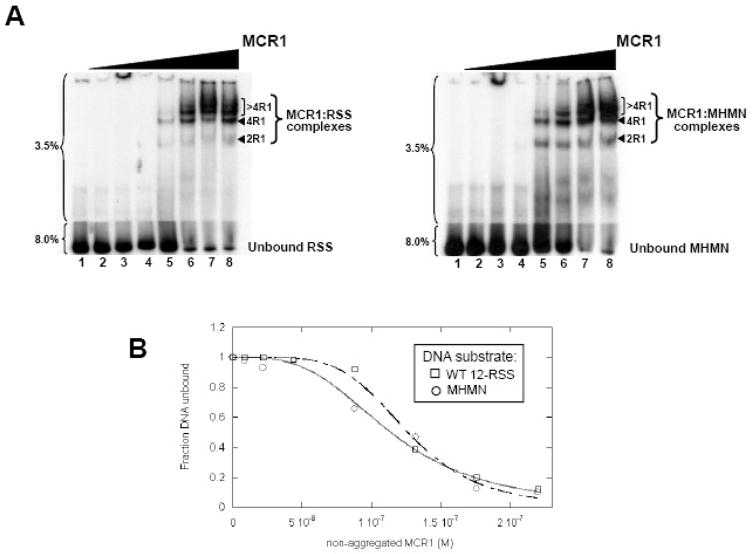
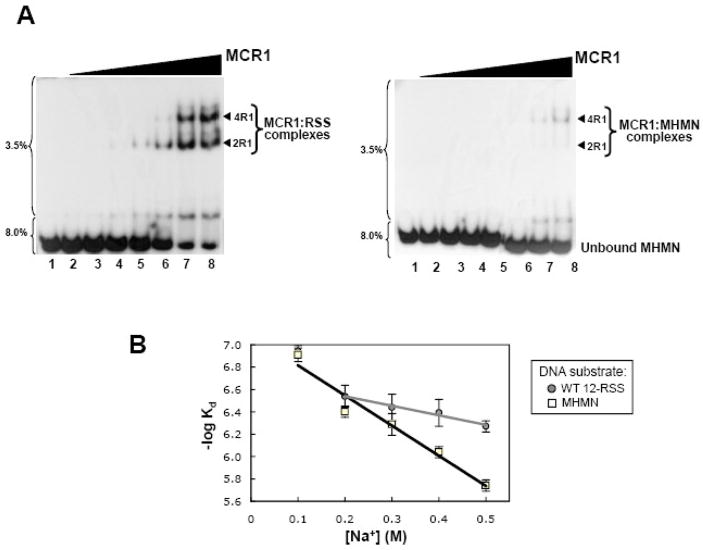
First, formation of the MCR1:oligonucleotide duplex complexes was monitored by EMSA using forward titration experiments (titration of MCR1 to each of the DNA substrates). MCR1 formed multiple complexes with both substrates in the forward titrations (Figure 1A), and the mobilities of the complexes are consistent with previous reports.14,26–28 The first shifted complex (labeled ‘2R1’ in Figure 1A) was previously shown to consist of a single MCR1 dimer bound to the oligonucleotide duplex.14,26–28 The next shifted complex (labeled ‘4R1’ in Figure 1A) was reported to contain two MCR1 dimers bound to the oligonucleotide duplex.27 Notably, based on pull-down assays (PD and KKR, unpublished results) and atomic force microscopy studies29, MCR1 dimers do not bridge between two separate oligonucleotide duplex molecules. Thus, the stoichiometries of the 2R1 and 4R1 complexes are at 1:1 and 2:1 MCR1 dimers per oligonucleotide duplex, respectively. Lastly, the slowest mobility complexes (labeled as ‘>4R1’ in Figure 1A) are not well-defined, and typically form only after the majority of the oligonucleotide substrate is saturated. Based on their mobilities, we presume that these complexes consist of additional dimers of MCR1 (3 or more) bound to the oligonucleotide duplex.27,28
Figure 1. MCR1 titration to WT 12-RSS versus MHMN as monitored by EMSA.
(A) EMSA of radiolabeled WT-12RSS (left panel) and MHMN (right panel) titrated with MCR1. The radiolabeled DNA substrates were titrated with increasing concentrations of MCR1 (lanes 2–8 in each panel contained 8.8, 22, 44, 88, 132, 175, and 220 nM MCR1, respectively). The binding buffer contained 0.1 M NaCl. Subsequent to a 30 min incubation at room temperature, the reactions were subjected to electrophoresis on a two-part 3.5/8.0% polyacrylamide gel. The percentage of polyacrylamide is labeled to the left of each gel. The MCR1:RSS complexes are labeled according to the stoichiometry of MCR1 bound to a single RSS duplex. For example, 2R1 and 4R1 consist of 2 subunits (or a dimer) and 4 subunits, respectively, of MCR1 bound to the RSS duplex.14,26–28 Complexes containing greater than 4 bound MCR1 subunits per RSS duplex are labeled >4R1. (B) Representative plots of the fraction of unbound WT 12-RSS (open squares) and unbound MHMN (open circles) with increasing MCR1 concentrations. Binding curves of MCR1 with WT 12-RSS (dashed line) and MHMN (solid line) were fit to equation 4 to obtain Kd values (Kdapp) and Hill coefficient values (n). From three independent experiments, Kdapp = 114±5 nM (n=3±1) and Kdapp = 123±7 nM (n=4±1) for the association of MCR1 with WT 12-RSS and MHMN substrate, respectively.
To construct binding curves to the forward titration data collected at 25°C, the MCR1 concentration was first corrected to account for protein aggregation (described in Supplemental Figure 1). The titration data were fit to equation 4 (see Materials and Methods), with the overall apparent Kd (Kdapp) values corresponding to the concentration of MCR1 at which 50% of the DNA substrate was bound and 50% remained unbound. For our purposes, using the overall Kdapp values was sufficient for comparing the interaction between MCR1 and the different DNA substrates. The resulting binding curves yielded Kdapp values of 114±5 nM and 123±7 nM for the association of MCR1 with WT 12-RSS and MHMN, respectively (Figure 1B). MCR1 bound cooperatively to both the WT 12-RSS and MHMN substrates, with Hill coefficients in the range of 3 to 4 (Figure 1B). (Hill coefficients were obtained from the fit to equation 4.) The Kdapp and Hill coefficient for the MCR1:WT 12-RSS interaction listed above are consistent with a previous report.14 (The binding affinity of MCR1 for a non-specific substrate has not been previously reported.) Overall, the forward titration results measured by EMSA showed that the affinity of MCR1 to the consensus WT 12-RSS versus the MHMN substrate were indistinguishable under these conditions (Figure 1).
A core RAG1 construct, expressed without the MBP fusion tag and referred to as tagless cR1, showed similar binding affinities to WT 12-RSS versus MHMN under the experimental conditions used in Figure 1 (Supplemental Figure 2). Moreover, as evident by EMSA, multiple protein-DNA complexes were formed (Supplemental Figure 2). Together these results demonstrate that the MBP portion of the MCR1 fusion protein did not affect the binding properties of core RAG1 to the DNA substrates in Figure 1. Compared to MCR1, tagless cR1 bound with significantly less affinity to the DNA substrates under the conditions used here (Supplemental Figure 2), possibly due to a relatively lower stability of tagless cR1. These results are not consistent with a previous study in which a significant difference in affinity between the tagless cR1 and WT 12-RSS versus MHMN substrates was reported.25 However, the previous study was conducted under different solution conditions (i.e. in 50 mM NaCl with no reducing agent), which may have yielded the disparity in results.
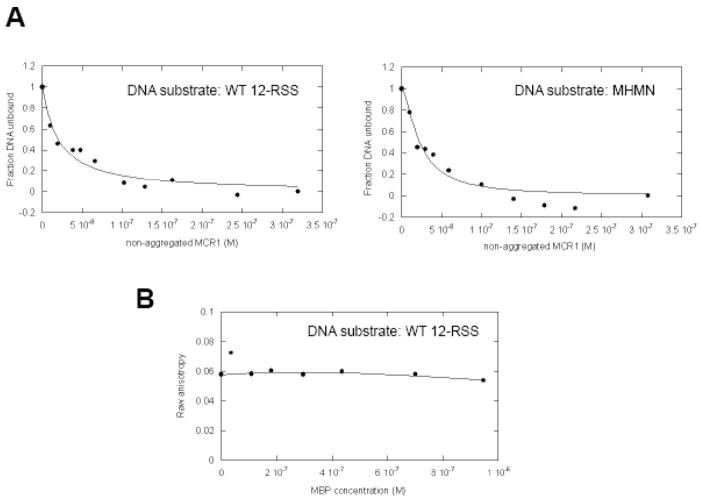
The relative binding affinities of MCR1 for the different DNA substrates were also monitored by fluorescence anisotropy. In these experiments, the WT 12-RSS and the MHMN substrates were labeled on the 5′ end of the top strand with Oregon Green 488. Oregon Green 488 is a useful probe for the investigation of protein-nucleic acid interactions by fluorescence anisotropy, given its fluorescence is photostable and is pH insensitive in the range from pH 6 to 9.30 Upon complex formation between protein and DNA, the fluorescence anisotropy of the probe is expected to increase due to reduced rotational mobility of the protein-DNA complex versus unbound DNA.31 Representative binding curves of the forward titration of MCR1 into each fluorescently labeled DNA substrate (Figure 2A) show that MCR1 bound to both the WT 12-RSS and the MHMN with similar affinities at 25°C, with Kdapp = 27±9 nM for WT 12-RSS versus 28±8 nM for the MHMN substrate. The discrepancy of the Kdapp values between fluorescence anisotropy and EMSA results can be attributed to the separation of bound complexes from free components during the electrophoresis step of the latter method, resulting in an underestimation of the binding affinities.31 The Hill coefficients also differ between the two methods (n~3 versus ~1 for the EMSA and fluorescence anisotropy results, respectively), which may also be due to the separation of components in the EMSA method. Nevertheless, both methods confirm that the binding affinity of MCR1 for DNA is sequence independent under the experimental conditions used.
Figure 2. MCR1 titrations to WT 12-RSS versus MHMN DNA as evaluated by fluorescence anisotropy methods.
(A) Equilibrium binding of MCR1 to Oregon Green-labeled WT 12-RSS (left panel) and MHMN (right panel) substrates. Shown are the representative binding curves fit to equation 1 (in Materials and Methods) for the titration (at 25°C) of MCR1 into WT 12-RSS (left panel) and MHMN (right panel), with each DNA substrate at 4 nM. Binding curves were fit to equation 1 to obtain Kd values (Kdapp) and Hill coefficient values (n). From three independent experiments, Kdapp = 22±6 nM (n=1.0±0.3) and Kdapp = 24±6 nM (n=1.5±0.5) for the association of MCR1 with WT 12-RSS and MHMN substrate, respectively. (B) Addition of MBP does not increase the anisotropy of Oregon Green labeled WT 12-RSS. The raw anisotropy data versus MBP concentration is shown. The titration was performed at 25°C in the same buffer as in panel A.
To confirm that the RAG1 region of the MCR1 fusion protein mediated the protein-DNA binding interaction, MBP alone was added to the Oregon Green labeled 12-RSS (Figure 2B). There was no detectable association between MBP and 12-RSS under these experimental conditions as shown by the constant anisotropy values over the course of the titration.
Non-specific DNA is an ineffective competitor for a pre-formed MCR1:WT 12-RSS complex
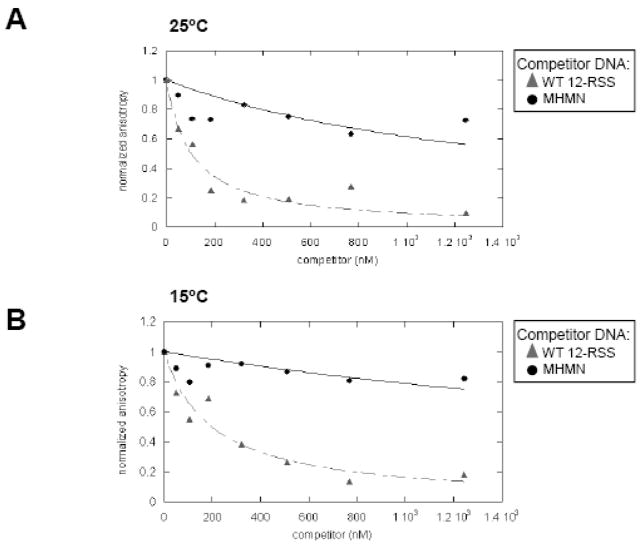
By the forward titration experiments, MCR1 bound equally well to the WT 12-RSS versus a non-specific DNA substrate (the MHMN substrate) under the conditions used in Figures 1&2. However, using similar conditions, previous results from EMSA competition assays showed a clear preference of MCR1 for the WT 12-RSS over the MHMN substrate.13,14 To confirm the previous EMSA results, competition assays were performed using fluorescence anisotropy methods. Increasing concentrations of non-labeled DNA (either WT 12-RSS or MHMN) were titrated to a pre-formed complex containing Oregon Green-labeled WT 12-RSS duplex (at 4 nM) and MCR1 (at 25 nM) at 25°C (Figure 3A). MCR1 concentrations used in the competition assays correspond to the midpoint of the titration curves in Figure 2. Therefore, the majority of the MCR1:Oregon Green-labeled WT 12-RSS complexes corresponded to 2R1 and 4R1 complexes (based on the EMSA results), since the higher order complexes (>4R1) were less apparent at the midpoint of the titration.
Figure 3. Competition assays of a pre-formed MCR1:WT 12-RSS complex as monitored by fluorescence anisotropy.
Representative plots depicting MCR1 (at 25 nM) complexed with Oregon Green-labeled WT 12-RSS (at 4 nM) titrated with unlabeled WT 12-RSS (filled triangles) or MHMN (filled circles) at 25°C (A) and 15°C (B). Equation 3 (see Materials and methods) was used to fit the competition data with the WT 12-RSS (dashed line) and MHMN (solid line) oligonucleotide duplexes as the competitors. In panel A, the value of Kc (derived from equation 3) with WT 12-RSS as competitor is 79 nM; the value of Kc with MHMN as competitor is estimated to be > 1200 nM. In panel B, Kc (with WT 12-RSS as competitor) is 150 nM, and Kc (with MHMN as competitor) is estimated to be > 2800 nM.
Under these conditions MCR1 demonstrated a substantially higher specificity for the WT 12-RSS than for the MHMN substrate, consistent with the previous results from EMSA competition assays.13,14 In addition, competition assays performed at 15°C (Figure 3B), with a higher fraction of the MCR1 sample in a non-aggregated form (Supplemental Figure 1), again demonstrated a sequence-specific interaction of MCR1 with the WT 12-RSS over the MHMN substrate.
MCR1 forms a longer-lived complex with WT 12-RSS versus non-specific DNA
The differing results from the forward titration versus the competition assays could be attributed to a separate non-specific DNA binding mode in MCR1 that masks any sequence-specific association with RSS in the forward titration experiments. Under the solution conditions used in Figures 1&2, the binding affinity of MCR1 for non-specific DNA may be similar to, or even greater than, that for the RSS bases in the WT 12-RSS substrate. We speculated that with two separate DNA binding modes (one sequence-specific and the other non-specific) the kinetics of the relative binding reactions would differ. For example, given the apparent specificity of MCR1 for WT 12-RSS, it is likely that the complex containing MCR1 bound to WT 12-RSS would display slower dissociation rates than MCR1 bound to MHMN. To compare the dissociation kinetics for the respective protein-DNA complexes, an excess of unlabeled DNA competitor (either WT 12-RSS or MHMN) was added to the pre-formed complexes of MCR1 with radiolabeled WT 12-RSS substrate. The dissociation of the protein from the radiolabeled substrate was monitored using EMSA (Figure 4), according to the protocol outlined previously by Gerstle and Fried.32
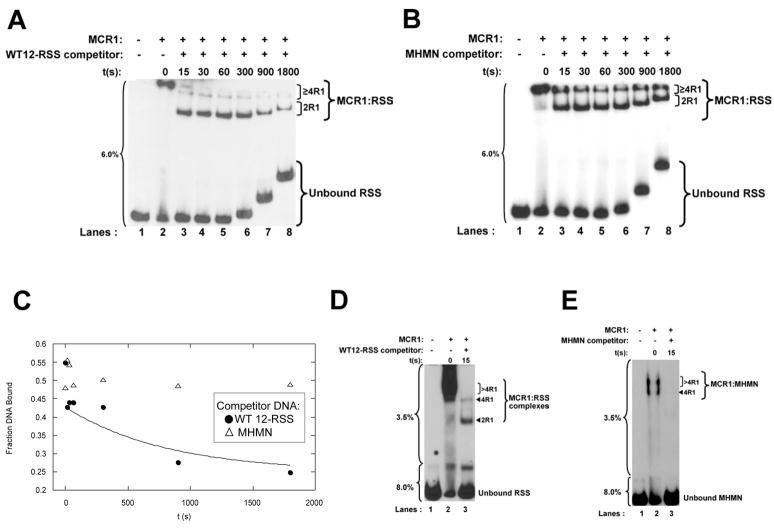
Figure 4. Kinetics of the dissociation of the MCR1:DNA complexes as a function of competitor DNA.
(A and B) Time course of the dissociation of MCR1 bound to radiolabeled WT 12-RSS upon addition of a 120-fold molar excess of unlabeled competitor DNA, with the competitor either (A) WT 12-RSS or (B) MHMN. At the indicated timepoints aliquots of the sample were loaded onto a single-part 6% nondenaturing polyacrylamide gel. The percentage of polyacrylamide is labeled to the left of each gel. The difference in migration position of equivalent bands between lanes arises from loading of the sample aliquots sequentially on a running gel. The MCR1:RSS complexes are labeled as 2R1 for dimeric MCR1 per RSS duplex, and ≥4R1 for complexes containing 4 or more MCR1 subunits per RSS duplex. The 4R1 and >4R1 complexes denoted in Figure 1A are not easily resolved on the 6% gels, and are thus grouped together. (C) Kinetics of the dissociation of the MCR1:WT12-RSS complex with WT 12-RSS (filled circles) and MHMN (open triangles) substrates as competitor. The time course data with the WT 12-RSS duplex as competitor was fit to equation 5 (using data from Panel A). The time point range from 15 sec (first time point value obtained after addition of competitor) to 1800 sec was used to obtain the koff value. The ‘Fraction DNA Bound’ (Y-axis label) corresponds to DNA bound in all MCR1:RSS complexes. (D) The first time point in panel A was repeated on a two-part 3.5%/8% gel (as labeled to the left of the gel) and the respective MCR1:RSS complexes labeled as in Figure 1A. (E) The MCR1:MHMN complexes are not detected 15 sec after addition of a 120-fold excess of unlabeled MHMN competitor. The samples were resolved on a two-part 3.5%/8% nondenaturing polyacrylamide gel as labeled to the left of the gel.
The protein-DNA complexes were resolved on a continuous 6% nondenaturing polyacrylamide gel (Figure 4A&B), which allowed dissociation kinetics to be monitored at longer time points while still retaining the protein-DNA complexes on the gel at the shorter time points. The initial protein-DNA complexes contained two, three, and possibly more, MCR1 dimers bound to the WT 12-RSS substrate (Figures 4A&B). Upon addition of either unlabeled WT 12-RSS or MHMN competitor, there was a rapid reduction (within 15 sec) in the stoichiometry of MCR1 bound to labeled WT 12-RSS from >4R1 at 0 sec to the appearance of 2R1 complex at 15 sec. (It is not possible to clearly distinguish 4R1 from >4R1 complexes on these gels.) This is presumably due to dissociation of MCR1 dimers bound nonspecifically to the WT 12-RSS substrate, while the specifically-bound MCR1 dimers are retained on the WT 12-RSS substrate (in the 2R1 and possibly 4R1 complexes). This is shown more clearly on a two-part, discontinuous (3.5%/8%) polyacrylamide gel (Figure 4D). Here, the protein-DNA complexes are separated at higher resolution, and the disappearance of the >4R1 complexes 15 sec after addition of competitor DNA is more easily visualized. Only the faster mobility complexes (2R1 and 4R1) remained after the initial time point.
Addition of unlabeled WT 12-RSS (at a 120-fold excess) disrupted the pre-formed MCR1 dimer:WT 12-RSS complex (2R1 complex) with a pseudo-first order rate constant (koff) of 0.0012±0.0001 sec−1 (from n=3 experiments), corresponding to a half life of 558±70 sec (Figure 4A&C). Very little dissociation of the MCR1:WT 12-RSS complexes were observed between the 15–1800 sec time points upon addition of an equivalent amount (a 120-fold excess) of unlabeled MHMN competitor (Figure 4B&C), again demonstrating the sequence specific interaction of MCR1 for the WT 12-RSS. In contrast to the results with the labeled WT 12-RSS substrate, complete dissociation of the pre-formed MCR1:radiolabeled MHMN complexes occurred within 15 sec after addition of a 120-fold excess unlabeled WT 12-RSS (not shown) or MHMN (Figure 4E), precluding determination of koff values.
Temperature dependence of MCR1 binding to specific versus non-specific DNA
We next asked how the relative affinities of MCR1 with the WT 12-RSS versus the MHMN DNA substrate would vary as a function of temperature. First, temperature dependent effects on the properties of the individual components were determined. The oligonucleotide duplexes remained in a stable double-stranded conformation over the temperature range in which the fluorescence anisotropy measurements were conducted based on melting curves (data not shown). MCR1, though, is known to form increasing amounts of inactive aggregates as the temperature approaches 37°C.28 Thus, the MCR1 concentration was corrected for protein aggregation at each temperature that the fluorescence anisotropy experiments were performed (Supplemental Figure 1).
Titrations of MCR1 to the Oregon Green labeled WT 12-RSS and MHMN DNA substrates were conducted over a range of temperatures between 4°C to 37°C and monitored by fluorescence anisotropy. Kdapp values were obtained by fitting the titration curves to equation 1 (Table 1). The Kdapp values for the interaction of MCR1 with the two DNA substrates were well within two-fold at each temperature monitored, with the exception of the titration data collected at 37°C. Only under this condition was there a >2-fold difference in Kdapp values (at 123 nM and 340 nM for the WT and MHMN substrates, respectively). Previous evidence suggests that both MCR1 and the RSS undergo conformational changes upon complex formation.27,29 Accordingly, the energetic barrier for conformational changes of MCR1 and/or the RSS may be more favorable at 37°C than at the lower temperatures, resulting in a greater differential between formation of the sequence-specific versus non-specific MCR1:DNA complexes.
Table 1.
Temperature dependence of MCR1 interaction with DNA substrates.
| WT 12-RSS |
MHMN |
|||
|---|---|---|---|---|
| Temp (K) | Kdapp (nM) | ΔG (kcal/mol)a | Kdapp (nM) | ΔG (kcal/mol)a |
| 278 | 61 ± 5 | −9.14 | 38 ± 9 | −9.40 |
| 283 | 32 ± 8 | −9.67 | 25 ± 6 | −9.81 |
| 288 | 48 ± 9 | −9.61 | 68 ± 9 | −9.41 |
| 293 | 49 ± 10 | −9.76 | 88 ± 10 | −9.42 |
| 298 | 27 ± 6 | −10.28 | 28 ± 8 | −10.26 |
| 303 | 80 ± 15 | −9.80 | 107 ± 11 | −9.63 |
| 310 | 123 ± 23 | −9.77 | 340 ± 57 | −9.14 |
ΔG was calculated using equation 2. Propagated errors on ΔG are all within ± 0.25 kcal/mol.
Ionic strength dependence of MCR1 binding to specific versus non-specific DNA
Since protein-non-specific DNA complex formations are dominated by electrostatic interactions, we next used EMSA to determine how the binding affinities of MCR1 with WT 12-RSS versus the MHMN DNA substrate varied with ionic strength (using buffers containing 0.1 to 0.5 M NaCl). The overall binding affinity decreased for the interaction of MCR1 with both DNA substrates upon an increase in NaCl concentration, as expected (Figure 5B). Notably, the binding affinity of MCR1 to MHMN was significantly reduced (particularly at 0.4–0.5 M NaCl) relative to the interaction with WT 12-RSS substrate. This difference at the higher NaCl concentrations is apparent in the EMSA results shown in Figure 5A, in which equivalent concentrations of MCR1 were titrated to each substrate at 0.4 M NaCl. At the highest MCR1 concentrations used in the titrations, only very low levels of the MCR1:MHMN complexes are formed (right panel) relative to MCR1:WT 12-RSS complex (left panel) in which the majority of DNA substrate was bound. Overall, these results indicate that electrostatic interactions play a greater role in stabilizing the non-specific (MCR1 with MHMN) versus the sequence-specific (MCR1 with WT 12-RSS) association. It is also apparent from the titration of MCR1 to WT 12-RSS (Figure 5A, left panel) that the higher order (>4R1) complexes are not evident at 0.4 M NaCl even with >90% of the WT 12-RSS substrate bound.
Figure 5. Ionic strength dependence on MCR1:DNA complex formation.
(A) Electrophoretic mobility shift assays of radiolabeled WT 12-RSS (left panel) and MHMN (right panel) titrated with MCR1 in binding buffer containing 0.4 M NaCl. The MCR1 concentration in lanes 2–8 in each panel was at 0.1, 0.2, 0.4, 0.6, 0.8, 1.2, and 1.6 μM, respectively. The MCR1:DNA complexes are labeled as in Figure 1A. (B) Plots of −log Kd versus NaCl concentration for the interaction of MCR1 with WT 12-RSS (gray circles) and MHMN (open squares). The error bars are from n=3 experiments. The linear least squares correlations are shown for the MCR1:MHMN association from 0.1–0.5 M NaCl (black line), and from 0.2–0.5 M NaCl for the MCR1:WT12-RSS association (gray line).
The plot of log Kd versus NaCl concentration for the MCR1:MHMN interaction shows a linear correlation over the entire NaCl concentration range measured (from 0.1M to 0.5M NaCl). However, the corresponding plot for the MCR1:WT 12-RSS interaction shows a linear correlation only over the 0.2–0.5M NaCl range, suggesting that the non-specific DNA binding mode is dominant at 0.1M NaCl. By extrapolation of the linear portion (0.2–0.5 M NaCl in Figure 5B) for the MCR1:WT 12-RSS interaction, we can estimate that at 0.1 M NaCl the Kdapp value is ~240 nM for the sequence-specific complex formation. Given that MCR1 associates with MHMN substrate at a Kdapp value of 123 nM at 0.1 M NaCl, then the relative fraction of WT 12-RSS that is complexed with MCR1 in a sequence-specific versus a non-specific binding mode can be estimated. For example, using the following expressions for Kd, the respective binding equilibrium expressions are:
where Kdapp(SP) and Kdapp(NSS) are the Kdapp values for the sequence-specific and non-specific complex formation (240 nM and 123 nM, respectively). Since Kdapp(SP)/Kdapp(NSS) is nearly 2, it follows that the ratio of non-specific complex to sequence-specific complex is ~2. Therefore, of the total amount of MCR1-bound WT 12-RSS substrate, ~1/3 is bound in a sequence-specific manner, and the remaining ~2/3 is bound via non-specific contacts only. This is likely a lower limit for the fraction of bound WT 12-RSS that is complexed by sequence-specific interactions, since it excludes 1) the possibility that an MCR1 dimer could simultaneously form the high affinity non-specific and sequence-specific contacts on the same oligonucleotide duplex molecule and 2) the cooperative binding of multiple MCR1 dimers to a single duplex.
Addition of RAG2 suppresses the interaction of RAG1 with non-specific DNA
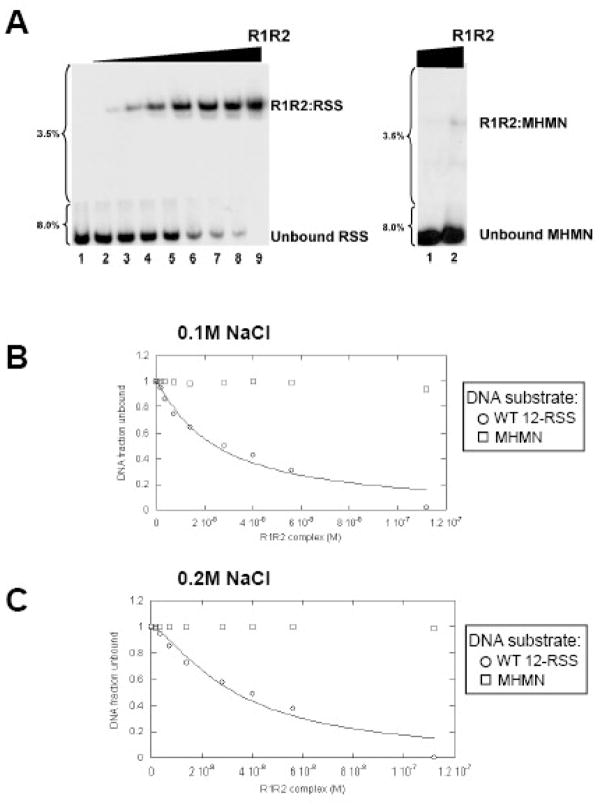
The RAG1:RAG2 complex, similarly to MCR1 alone, binds to a single DNA substrate. Only in the presence of HMGB1 or HMGB2 can the RAG1:RAG2 complex specifically bridge a 12-RSS with a 23-RSS substrate to form the synaptic complex.33,34 To examine the effects of RAG2 on the DNA-binding property of RAG1, the binding affinities of the MCR1:GST-core RAG2 complex (termed R1R2 complex) with WT 12-RSS and MHMN substrates were compared by EMSA. Direct titration of the R1R2 complex (co-expressed and purified from 293T cells) into solutions containing either the 32P-labeled WT 12-RSS or the MHMN substrate yielded one major protein-DNA complex (in binding buffer containing 0.1 M NaCl) (Figure 6A). The protein-DNA complex consists of two subunits of each RAG protein, as described previously.28 The binding affinity of the R1R2 complex is markedly higher for WT 12-RSS than for MHMN substrate, with the Kdapp value for the interaction with WT 12-RSS complex at 25±5 nM as derived from equation 4 (Figure 6B). In contrast, only a small fraction (~1%) of the 32P-labeled MHMN substrate was bound by the highest concentrations of the R1R2 complex (Figure 6A). We estimate the Kd>10 μM for this latter interaction, assuming a similar Hill coefficient as shown with the WT 12-RSS substrate (at n=1.0±0.3; see Figure 6 legend). Similar results were obtained using RAG proteins that were individually expressed and purified from different sources (MCR1 expressed in E. coli and GST-core RAG2 expressed in 293T cells), and then mixed prior to the addition of the RSS substrates (Supplemental Figure 3). Specifically, in binding buffers containing 0.1 M NaCl, the reconstituted R1R2 complex bound the WT 12-RSS substrate with Kd = 87±10 nM, whereas an interaction with the MHMN substrate was not observed at the highest concentrations of reconstituted R1R2 complex used (224 nM). While the binding affinity to WT 12-RSS was somewhat less with the reconstituted versus the co-expressed R1R2 complex, these results demonstrate that the differential affinity between the RSS and non-RSS substrates did not require co-folding of the RAG proteins or expression of MCR1 in mammalian cells.
Figure 6. Titration of the R1R2 complex into WT 12-RSS versus MHMN substrate as monitored by EMSA.
(A) Increasing concentrations of the R1R2 complex were titrated into either WT 12-RSS (left panel) or MHMN (right panel). R1 is MCR1 and R2 is GST-core RAG2 co-expressed and purified from 293T cells. The binding reactions were performed in buffer containing 0.1M NaCl. The concentration of the R1R2 complex (in terms of MCR1) in lanes 2–9 in the left panel was at 1.8, 3.5, 7, 14, 28, 40, 56, and 112 nM, respectively. Only the lanes containing 56nM (lane 1) and 112 nM (lane 2) of R1R2 complex concentrations are shown in the right panel, as protein-DNA complexes were not detected at lower concentrations. (B & C) Representative plots of the fraction of WT 12-RSS unbound (open circles) and MHMN unbound (open squares) with increasing concentrations of the R1R2 complex. The binding curves for the interaction of the R1R2 complex with the WT-12-RSS substrate (solid line) at either 0.1M NaCl (panel B) or 0.2 M NaCl (panel C) were fit to equation 4 as described in Materials and Methods. From three independent experiments, association of the R1R2 complex with WT 12-RSS yielded Kdapp = 25±5 nM (n=1.0±0.3) and Kdapp = 32±5 nM (n=1.5±0.3) at 0.1 M and 0.2 M NaCl, respectively.
Determination of binding affinities for the R1R2 complex with the Oregon Green-labeled DNA substrates could not be accurately determined by fluorescence anisotropy since GST-core RAG2 appeared to interact with the Oregon Green fluorophore, as well as other fluorophores tested (including fluorescein and Alexa Fluor 488; data not shown). Nevertheless, direct titrations monitored by this method did show a differential affinity of the RAG complex to the WT 12-RSS versus the MHMN DNA substrate (data not shown), although not to the extent as seen in the EMSA results, which may be due to the GST-core RAG2:fluorophore interaction.
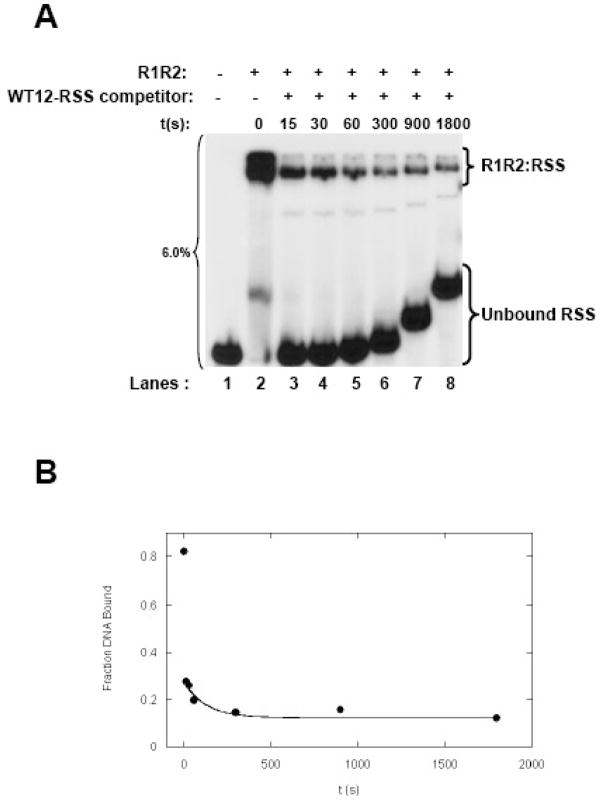
The pseudo-first order koff value for the dissociation of the preformed R1R2:WT 12-RSS complex by a 120-fold excess of unlabeled WT 12-RSS was at 0.0096±0.002 sec−1 (from n=3 experiments), corresponding to a half life of 72±22 sec (Figure 7). It was somewhat unexpected that the R1R2 complex showed a shorter residence time on WT 12-RSS, as compared to MCR1 in the absence of R2. However, it may be that the R1R2 complex is in a conformation primed for sequence-specific interactions with the RSS, unlike MCR1 alone. Since MCR1 (in the absence of R2) may undergo conformational changes upon dissociating from the RSS,27 there may be energetic barriers that impede its dissociation relative to the R1R2:WT12-RSS complex. That the R1R2 complex can readily dissociate from the RSS is consistent with previous results inferred from kinetic studies on the RSS nicking reaction,35 but is in contrast with another study that concluded the R1R2:RSS complex did not dissociate after a 1-hour time period even in the presence of a 1000-fold excess of unlabeled RSS.13 However, the protein-DNA complexes were crosslinked prior to EMSA in the latter study, which may have led to the differing results.
Figure 7. Kinetics of the dissociation of the R1R2:WT 12-RSS as a function of competitor DNA.
(A) Time course of the dissociation of reconstituted R1R2 complex bound to radiolabeled WT 12-RSS upon addition of a 120-fold molar excess of unlabeled WT 12-RSS competitor. At the indicated times, aliquots of the sample were loaded onto a 6% nondenaturing polyacrylamide gel. The difference in migration position of equivalent bands between lanes arises from loading of the sample aliquots sequentially on a running gel. (B) Kinetics of the dissociation of the reconstituted R1R2:WT 12-RSS complex. Representative plot of fraction DNA bound versus time was fit using data from Panel A as described in Figure 4.
Overall, the largest impact upon addition of RAG2 to the titration experiments at 0.1M NaCl was a substantial decrease in the interaction with the MHMN substrate. For example, a comparison of the EMSA results show that MCR1 (at ~100 nM) half-saturated the MHMN DNA substrate (Figure 1B), whereas an equivalent concentration of R1R2 formed only minute amounts of complex with the DNA substrate (Figure 6A, lane 2 in right panel; and Supplemental Figure 3, lane 2 in right panel). The binding affinities of the R1R2 complex versus MCR1 alone for the non-specific DNA substrate differed by more than two orders of magnitude (Kdapp > 10 uM for the interaction of the MHMN substrate with the R1R2 complex versus Kdapp = 123 nM with MCR1), corresponding to a ΔΔG of >2.7 kcal/mol.
It should be noted that titration experiments using MCR1 in the absence, but not in the presence, of GST-core RAG2 were corrected for protein aggregation (Supplemental Figure 1). This is since addition of RAG2 appeared to reduce the non-specific aggregation of MCR1, and thereby increased the DNA binding activity of MCR1 at 25°C (Supplemental Figure 4). Even so, applying the correction factor reduced the Kd value for the MCR1:MHMN interaction by only ~2-fold at 25°C (Supplemental Figure 1), and therefore has no impact on the conclusion that the addition of RAG2 substantially weakens the interaction of MCR1 with non-specific DNA.
The interaction of the R1R2 complex with the WT 12-RSS substrate was resistant to moderate changes in ionic strength, with a Kd value of 32±5 nM in binding buffer containing 0.2 M NaCl (Figure 6C), demonstrating that the sequence-specific complex is not driven by electrostatic interactions. Under the same conditions (at 0.2 M NaCl), association of the R1R2 complex with the MHMN substrate was not evident (Figure 6C).
DISCUSSION
The initial step in V(D)J recombination is the sequence-specific recognition of an RSS by the RAG proteins; however the relevant role of each RAG protein towards this recognition event is not clear. Here, using a combination of EMSA and fluorescence anisotropy methods, we have gained insight into the both the sequence specific versus non-specific DNA binding modes of core RAG1. In this study, core RAG1 is fused to the C-terminal end of maltose binding protein, and is referred to as MCR1. Our results also show that RAG2 can modulate the non-specific DNA binding mode of MCR1 at physiological conditions.
Notably, in the absence of RAG2, MCR1 exhibited similar overall binding affinities to the WT 12-RSS substrate and a non-specific DNA substrate (MHMN) at physiological ionic strengths (0.1 to 0.2 M NaCl). However, an increase of ionic strength to 0.4 to 0.5 M NaCl yielded a more energetically favorable association of MCR1 with the WT 12-RSS versus the MHMN substrate. Even though the overall binding affinities are similar at 0.1 M NaCl, it is likely the binding kinetics differ for formation of the respective complexes. For example, proteins complexed with non-specific DNA typically demonstrate relatively rapid on and off binding rates, since the complexes are primarily stabilized by electrostatic contacts with little or no conformational changes in either protein or DNA component.36 In contrast, sequence-specific interactions often involve conformational changes of protein or DNA upon formation of base-specific contacts, which can result in relatively slower on and off rates.36 As a result, formations of sequence-specific protein-DNA complexes are often facilitated by initially forming contacts in a non-specific manner. Here, results from the time course competition assays suggest a similar effect in that the dissociation kinetics for the MCR1:WT 12-RSS complex was notably slower than for the MCR1:MHMN complex. It is also likely that the on rate for the formation of the MCR1:WT 12-RSS complex is slower than formation of the MCR1:MHMN complex, as both MCR1 and the WT 12-RSS undergo conformational changes upon complex formation.27,29
Despite similar overall binding affinities to the two DNA substrates at 0.1 M NaCl, MCR1 clearly recognized the WT 12-RSS over the MHMN substrate. Specifically, both the steady state and time course competition assays (Figures 3&4) showed that the MCR1:MHMN complex can be displaced by either DNA substrate. In contrast, the MCR1:WT 12-RSS complex is not readily displaced by the MHMN substrate. Overall, in buffers containing 0.1 M NaCl, the forward titrations indicated that MCR1 and WT 12-RSS associate in a non-specific manner, whereas competition assays showed the formation of sequence-specific complexes. How can these apparently contradictory results be resolved? Based on the ionic strength dependence of the MCR1:DNA substrate binding affinities (Figure 5), we estimate that a significant fraction of the WT 12-RSS binds MCR1 via base-specific contacts at 0.1 M NaCl. Specifically, the minimum fraction of complex formed through sequence-specific contacts is ~1/3 of the total bound DNA substrate. This is a substantial fraction of the complex that would demonstrate preferential substrate binding in the competition assays. It is possible that the fraction of sequence-specifically bound substrate is larger than 1/3, since association of a MCR1:WT 12-RSS complex through non-specific contacts could subsequently form sequence-specific contacts on the same DNA molecule.
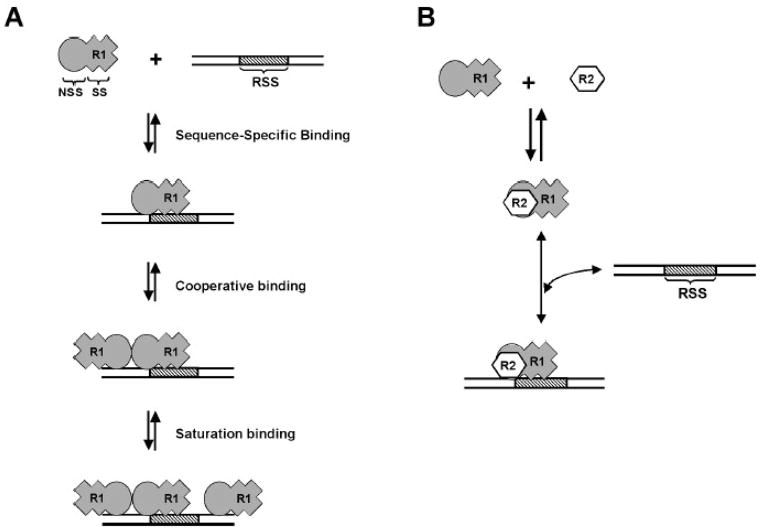
MCR1 forms multiple complexes with the DNA substrates, with two protein-DNA complexes of differing stoichiometries apparent (labeled 2R1 and 4R1 in Figure 1). Both the 2R1 and 4R1 complexes demonstrate specificity for the WT 12-RSS substrate in the time course dissociation experiments (Figure 4B). Moreover, the levels of 4R1 complex were decreased to a greater extent than the 2R1 complex upon addition of excess WT 12-RSS competitor, indicating that the 2R1 complex may be the more stable complex (Figure 4B). Based on these results, we propose the following model for the interaction of MCR1 with the WT 12-RSS substrate. First, a single MCR1 dimer binds the WT 12-RSS and forms base-specific contacts with the 12-RSS (Figure 8A), which is followed by the binding of a second MCR1 dimer. Binding of the second MCR1 dimer to form the 4R1 complex may involve interdimer contacts on the DNA substrate. As a result, although the second MCR1 dimer to associate with the complex may not form direct contacts with the RSS, the resulting 4R1 complex will show sequence-specificity based on the interdimer contacts (Figure 8A). It is likely that any additional MCR1 dimers that bind to the DNA substrate do so at sequences distal to the RSS site (Figure 8A), since competitor DNA more easily displaced these complexes. Similar complexes as shown in Figure 8A may form between MCR1 and the MHMN substrate, since complexes of the same stoichiometries formed between MCR1 and the MHMN substrate versus the WT 12-RSS substrate (Figure 1A). However, the protein-DNA contacts with the MHMN substrate would be primarily through the non-specific DNA binding mode of MCR1.
Figure 8.
Schematic model for the interaction of RAG1 alone and the RAG1:RAG2 complex with the RSS. (A) Dimeric core RAG1 (R1) contains sequence-specific (SS) and non-specific (NSS) DNA binding sites. In the first binding event shown, R1 forms sequence-specific contacts with the RSS through the SS DNA binding sites (corresponds to the ‘2R1’ MCR1:RSS complex in Fig. 1). Although not represented here, dimeric R1 can also bend the RSS substrate.29 In the next binding event shown, a second R1 dimer binds cooperatively forming protein-protein contacts with the sequence-specific bound R1 dimer (corresponds to the ‘4R1’ complex in Fig. 1). In the last binding event shown, additional R1 dimers may bind to the DNA substrate with high concentrations of protein (corresponds to ‘>4R1’ complexes in Fig. 1). (B) Complex formation of R1 with core RAG2 (R2) allows sequence-specific complex formation with the RSS, but blocks the high-affinity NSS DNA interactions, and the cooperative binding of a second R1 dimer.
In contrast to MCR1 alone, the MCR1:GST-core RAG2 (R1R2) complex bound with significantly reduced affinity to non-specific DNA, suggesting that the non-specific DNA binding mode of MCR1 is suppressed by RAG2. Overall, this would result in a marked increase in the binding differential between the RSS and non-specific DNA, thereby increasing the specificity of DNA binding (Figure 8B). Furthermore, complex formation of MCR1 with GST-core RAG2 prevented association with additional MCR1 dimers on the DNA substrate, since only a single complex of protein-DNA complex was observed in the titrations of R1R2 to the WT 12-RSS substrate.
By measuring binding kinetics, we determined that the R1R2 complex can readily dissociate from WT 12-RSS upon addition of an RSS competitor. These results suggest that within developing lymphocytes, it is possible the R1R2 complex could sample multiple RSS sites along the appropriate antigen receptor loci prior to initiating the nicking reaction. However, subsequent to the nicking reaction, it has been shown that the RAG proteins do not readily dissociate from the nicked RSS.37 Possibly, the nicking reaction induces additional conformational changes in the DNA, such as base unpairing, that produce enhanced interactions with the RAG proteins. If so, the R1R2 complex bound to a nicked RSS would then be primed to carry out subsequent steps, including associating with a partner 23-RSS.
Other sequence-specific DNA-binding proteins that show a non-specific DNA-binding mode includes p5338,39 and the E1 initiator protein from bovine papillomavirus.40 In both proteins a non-specific DNA-binding domain effectively suppressed the interaction of the specific-binding domain with the recognition site. For the E1 initiator protein, the suppression of the sequence-specific interaction can be overcome by association with the virus-encoded transcription factor E2, which blocks binding of DNA to the non-specific DNA-binding domain.40 Likewise, the binding ability of the non-specific DNA-binding domain in p53 was shown to be blocked by either association with several different proteins or by post-translational modification of the non-specific DNA-binding domain.39 By either mechanism, reduction of the non-specific DNA-binding function of p53 removed the suppression of the sequence-specific DNA-binding domain, thereby showing an apparent increase in binding specificity.
There are several regions in core RAG that have been shown to interact with non-specific DNA. These include the NBR and the C-terminal domain, both of which contain multiple regions rich in basic residues.41 While recognition and binding to the RSS is a prerequisite for RAG-mediated site-specific cleavage, binding to DNA in a sequence-independent manner is also important at several steps in the V(D)J recombination process. First, RAG contacts with RSS-flanking nucleotides (i.e. in the V, D, or J coding regions) may assist in orienting the RAG active site at the coding flank/RSS heptamer border.17,42 Second, interaction with the flanking nucleotides is also necessary for hairpin formation at the coding end.43,44 Third, the RAG proteins have been shown to maintain contact with the coding end hairpins after DNA cleavage, which may be a crucial step in the transfer of DNA DSB products to the NHEJ DNA repair pathway.45,46 In all of these steps, it is possible that RAG2 modulates the non-specific DNA binding activity of core RAG1 to only associate with non-specific DNA in the appropriate context during V(D)J recombination. Alternatively, certain interactions of the R1R2 complex with non-specific DNA would be potentially harmful. For example, the RAG proteins have been shown to catalyze tranposition events in vitro and, at very low levels, in vivo.47,48 The strand transfer step in the transposition reaction involves association of the R1R2 complex with target DNA in a non-specific manner. The regulation of the non-specific DNA binding mode in RAG1, by RAG2, may be necessary to prevent such deleterious events during lymphocyte development. Further studies into the role of non-specific DNA interactions during V(D)J recombination, and the regulation of these interactions, will be important for a greater understanding of this intricate process.
MATERIALS AND METHODS
Protein expression and purification
The plasmid pCJM233 (described previously)14 containing the gene encoding for core RAG 1 fused to the C-terminal end of maltose binding protein (MBP) was transformed into Escherichia coli BL21 cells. The MBP-core RAG1 fusion protein consisted of 384–1008, termed MCR1, was expressed and purified as previously described.17 The concentration of active MCR1 purified from Escherichia coli was determined by UV absorbance at 280 nm using an extinction coefficient of 129.5 mM−1 cm−1. In experiments with MCR1 only (in the absence of GST-core RAG2), the MCR1 concentration was subsequently corrected for aggregation as shown in Supplemental Figure 1.
The protein, MBP, was collected from the final size exclusion chromatography (SEC) purification step of MCR1, and corresponds to the ~10% of MCR1 that is degraded (to MBP) upon expression and purification. The concentration of MBP was determined by UV absorbance at 280 nm using an extinction coefficient of 66.5 mM−1 cm−1.
GST-core RAG2 consists of residues 1–387 of the full-length RAG2 fused to the C-terminal end of glutathione S-transferase (GST). GST-core RAG2, either alone or with MCR1, was transiently expressed and purified from 293T cells as previously described.49 The concentration of the RAG proteins expressed and purified from 293T cells were determined by Western blotting using monoclonal anti-GST or anti-MBP antibodies as previously described.49 The concentration of the co-expressed MCR1:GST-core RAG2 complex (referred to as the R1R2 complex) is given in terms of the determined MCR1 concentration.
Oligonucleotide substrates
The sequence of the top strand of wild-type 12-RSS (WT 12-RSS) is 5′-GATATGGCTCGTCTTACACAGTGATATAGACCTTAACAAAAACCTCCAATCGAGCGGAG-3′. The sequence of the top strand mutant heptamer and mutant nonamer DNA substrate (MHMN) is identical to the wild-type RSS except the heptamer (CACAGTG) and the nonamer (ACAAAAACC) are replaced by (GAGAAGC) and (AGGCTCTGA), respectively. In the fluorescence experiment, both the top strands of WT 12-RSS and MHMN were conjugated to the fluorophore Oregon Green 488 at the 5′ end (commercially synthesized and PAGE purified by Integrated DNA Technologies). Double-stranded DNA substrates were prepared by annealing the fluorescently-labeled top strand WT 12-RSS or MHMN with their respective complement.
Fluorescence anisotropy assay
Oregon Green-labeled WT 12-RSS or MHMN duplex (at 4 nM) was titrated with increasing concentrations of MCR1. The binding buffer contained 10 mM Tris pH 8.0; 5mM MgCl2; 100mM NaCl; 2mM dithiothreitol (DTT). Protein-DNA interactions were monitored by the anisotropy signal change as the protein-DNA complex was formed. Fluorescence anisotropy experiments were performed using an SLM-800 spectrofluorometer equipped with 450W Xenon lamp. The excitation and emission wavelength for the Oregon green fluorophore were set at 494 and 518 nm, respectively. For each titration, the sample was incubated at the indicated temperature for 8 minutes to reach equilibrium, after which the anisotropy was measured at least 10 times. G-factor corrections were included in all calculated anisotropy values. Anisotropy values were calculated using the program VINCI (ISS, Champaign, IL). The temperature range was from 5°C to 37°C.
In fluorescence competition assays, Oregon Green-labeled WT 12-RSS (at 4 nM) was incubated with MCR1 (at 25 nM). After equilibrium was reached in formation of the protein-DNA complex (no further change observed in anisotropy values), either nonlabeled WT 12-RSS or MHMN was added to the reaction at increasing concentrations. The reactions were performed in the binding buffer containing 0.1 M NaCl (at 15 or 25°C) as described above. Prior to recording anisotropy values, the samples were incubated 10 min after each addition of nonlabeled competitor, as the anisotropy values did not significantly change (<5%) with longer incubation times.
Fluorescence data analysis
Binding curves for the interaction of MCR1 and the DNA substrates obtained from fluorescence anisotropy were fit to the following binding isotherm:
| (1) |
where ΔA is the normalized change in fluorescence anisotropy with 1−ΔA corresponding to the fraction of unbound DNA; P is the concentration of MCR1, n is the Hill coefficient, and Kd is the dissociation constant. Kd values reported are the average from n=3 independent experiments. Curve fitting was performed using the program KaleidaGraph (Synergy Software). The ΔG at each temperature was calculated from its corresponding Kd (calculated from equation 1) as follows:
| (2) |
where ΔG is the free energy of binding, R is the universal gas constant, and T is the temperature in Kelvin.
Curve fitting to the results from the competition assays (monitored by fluorescence anisotropy) were done using nonlinear least squares fits according to the following equation:
| (3) |
where A0 is the fluorescence anisotropy value of free Oregon-labeled WT 12-RSS; A+ and A− are the fluorescence anisotropy value in the presence and absence of competitor DNA, respectively; PT is the total protein concentration and CT is the total concentration of unlabeled competitor. In addition, Kc corresponds to the concentration of competitor DNA (either WT 12-RSS or MHMN) required to reduce the anisotropy value by 50%.
Electrophoretic mobility shift assay (EMSA) – Measurement of binding affinities
Oligonucleotide duplexes each containing a WT 12-RSS or a MHMN sequence were used as DNA substrates in the EMSA experiments. The sequence of the WT 12-RSS and the MHMN are the same as used in the fluorescence anisotropy measurements, but without the Oregon Green fluorescent label. The WT 12-RSS and MHMN substrates were labeled with 32P at the 5′-end of the top strand using γ32P-ATP and T4 polynucleotide kinase. The double-stranded substrates were prepared by annealing the top strands with their respective complementary oligonucleotides.
EMSA was performed as described previously.27 The binding buffer contained 20 mM Tris, pH 8.0; 5 mM MgCl2; 6% glycerol; and 2 mM dithiothreitol (DTT). The NaCl concentration ranged from 0.1 to 0.5 M, as specified. Samples were incubated for 30 minutes at room temperature, and the protein-DNA complexes subsequently resolved on a two-part 3.5/8% nondenaturing polyacrylamide gel. Gels were dried and quantitated using an Amersham Biosciences SI phosphorImager and densitometer. The fraction of unbound DNA versus protein concentration was plotted and the data points were fit to the following equation:
| (4) |
where Kd is the dissociation constant, P is the protein concentration and n is the Hill coefficient.14 Kd values reported are the average from n=3 independent experiments.
Electrophoretic mobility shift assay (EMSA) – Measurement of binding kinetics
For the time course competition experiments, MCR1 (at 132 nM) or reconstituted R1R2 complex (with R1 concentration at 150nM, and the R2 concentration at 75nM) was incubated with 1nM 32P-labeled WT 12-RSS (or in some cases 1 nM 32P-labeled MHMN substrate as indicated) at 25°C for 1 hour. The binding reactions were performed under the same conditions as described in the previous section. Subsequently, 120-fold excess of competitor unlabeled DNA (either WT 12-RSS or MHMN DNA as indicated) was added to the sample reaction. After different incubation times, the reactions were subjected to electrophoresis on a single-part 6% nondenaturing polyacrylamide gel, unless otherwise stated. The fraction of bound DNA at each time point was quantitated and plotted versus time. The data points were fit to the single exponential according to the following equation:
| (5) |
Where Y0 is the binding at time zero; NS is the binding at infinite time; k is the rate constant in inverse unit of time. The k values reported (koff) are the average from n=3 independent experiments.
Acknowledgments
We thank Jialing Lin for assistance with the fluorescence anisotropy measurements, Bruce Baggenstoss for assistance with MALLS-SEC, and Ruby Rahman for assistance with protein purification. This work was supported by National Institutes of Health grant AI-054467 and an Oklahoma Center for Advancement in Science and Technology award (HR05-101) to KKR. LMG was supported by National Institutes of Health award F32 AI069784.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 2.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 3.West RB, Lieber MR. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol Cell Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mundy CL, Patenge N, Matthews AGW, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–24. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–33. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 8.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 9.Eastman QM, Schatz DG. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu K, Lieber MR. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol Cell Biol. 2000;20:7914–7921. doi: 10.1128/mcb.20.21.7914-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–9. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 12.Nagawa F, Ishiguro K-I, Tsuboi A, Yoshida T, Ishikawa A, Takemori T, Otsuka AJ, Sakano H. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol Cell Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodgers KK, Villey IJ, Ptaszek L, Corbett E, Schatz DG, Coleman JE. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 16.Spanopoulou E, Zaitseva F, Wang FH, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 17.Arbuckle JL, Fauss LJ, Simpson R, Ptaszek LM, Rodgers KK. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J Biol Chem. 2001;276:37093–37101. doi: 10.1074/jbc.M105988200. [DOI] [PubMed] [Google Scholar]
- 18.Peak MM, Arbuckle JL, Rodgers KK. The central domain of core RAG1 preferentially recognizes single-stranded recombination signal sequence heptamer. J Biol Chem. 2003;278:18235–18240. doi: 10.1074/jbc.M302041200. [DOI] [PubMed] [Google Scholar]
- 19.De P, Rodgers KK. Putting the pieces together: Identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol Rev. 2004;200:70–82. doi: 10.1111/j.0105-2896.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 20.Cuomo CA, Mundy CL, Oettinger MA. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson PC, Desiderio S. V(D)J recombination signal recognition: Distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 23.Nishihara T, Nagawa F, Imai T, Sakano H. RAG-heptamer interaction in the synaptic complex is a crucial biochemical checkpoint for the 12/23 recombination rule. J Biol Chem. 2008;283:4877–4885. doi: 10.1074/jbc.M709890200. [DOI] [PubMed] [Google Scholar]
- 24.Mo X, Bailin T, Sadofsky MJ. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J Biol Chem. 1999;274:7025–7031. doi: 10.1074/jbc.274.11.7025. [DOI] [PubMed] [Google Scholar]
- 25.Ciubotaru M, Ptaszek LM, Baker GA, Baker SN, Bright FV, Schatz DG. RAG1-DNA binding in V(D)J recombination: Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J Biol Chem. 2003;278:5584–5596. doi: 10.1074/jbc.M209758200. [DOI] [PubMed] [Google Scholar]
- 26.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godderz LJ, Rahman NS, Risinger GM, Arbuckle JL, Rodgers KK. Self-association and conformational properties of RAG1: Implications for formation of the V(D)J recombinase. Nucleic Acids Res. 2003;31:2014–2023. doi: 10.1093/nar/gkg281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De P, Zhao S, Gwyn LM, Godderz LJ, Peak MM, Rodgers KK. Thermal dependency of RAG1 self-association properties. BMC Biochem. 2008;9:5. doi: 10.1186/1471-2091-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavlicek JW, Lyubchenko YL, Chang Y. Quantitative analyses of RAG-RSS interactions and conformations revealed by atomic force microscopy. Biochemistry. 2008;47:11204–11. doi: 10.1021/bi801426x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusinova E, Tretyachenko-Ladokhina V, Vele OE, Senear DF, Alexander Ross JB. Alexa and Oregon Green dyes as fluorescence anisotropy probes for measuring protein-protein and protein-nucleic acid interactions. Anal Biochem. 2002;308:18–25. doi: 10.1016/s0003-2697(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 31.LiCata VJ, Wowor AJ. Applications of fluorescence anisotropy to the study of protein-DNA interactions. Methods Cell Biol. 2008;84:243–62. doi: 10.1016/S0091-679X(07)84009-X. [DOI] [PubMed] [Google Scholar]
- 32.Gerstle JT, Fried MG. Measurement of binding kinetics using the gel electrophoresis mobility shift assay. Electrophoresis. 1993;14:725–31. doi: 10.1002/elps.11501401115. [DOI] [PubMed] [Google Scholar]
- 33.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 34.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 35.Yu K, Taghva A, Ma Y, Lieber MR. Kinetic analysis of the nicking and hairpin formation steps in V(D)J recombination. DNA Repair (Amst) 2004;3:67–75. doi: 10.1016/j.dnarep.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–35. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grawunder U, Lieber MR. A complex of RAG1 and RAG2 proteins persists on DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res. 1997;25:1375–1382. doi: 10.1093/nar/25.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayle JH, Elenbaas B, Levine AJ. The carboxyl-terminal domain of the p53 protein regulates sequence-specific DNA binding through its nonspecific nucleic acid-binding activity. Proc Natl Acad Sci U S A. 1995;92:5729–33. doi: 10.1073/pnas.92.12.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Kulesz-Martin MF. Sliding into home: facilitated p53 search for targets by the basic DNA binding domain. Cell Death Differ. 2006;13:881–4. doi: 10.1038/sj.cdd.4401905. [DOI] [PubMed] [Google Scholar]
- 40.Stenlund A. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. Embo J. 2003;22:954–63. doi: 10.1093/emboj/cdg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spanopoulou E, Cortes P, Shih C, Huang CM, Silver DP, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 42.Mo X, Bailin T, Sadofsky MJ. A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol Cell Biol. 2001;21:2038–2047. doi: 10.1128/MCB.21.6.2038-2047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat Struct Mol Biol. 2006;13:1010–5. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 44.Grundy GJ, Hesse JE, Gellert M. Requirements for DNA hairpin formation by RAG1/2. Proc Natl Acad Sci U S A. 2007;104:3078–83. doi: 10.1073/pnas.0611293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 46.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–6. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 47.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 48.Brandt VL, Roth DB. V(D)J recombination: how to tame a transposase. Immunol Rev. 2004;200:249–60. doi: 10.1111/j.0105-2896.2004.00161.x. [DOI] [PubMed] [Google Scholar]
- 49.Rahman NS, Godderz LJ, Stray SJ, Capra JD, Rodgers KK. DNA cleavage of a cryptic recombination signal sequence by RAG1 and RAG2: Implications for partial VH gene replacement. J Biol Chem. 2006;281:12370–12380. doi: 10.1074/jbc.M507906200. [DOI] [PubMed] [Google Scholar]