Abstract
Characterizing how genomic sequence interacts with trans-acting regulatory factors to implement a program of gene expression in eukaryotic organisms is critical to understanding genome function. One means by which patterns of gene expression are achieved is through the differential packaging of DNA into distinct types of chromatin. While chromatin state exerts a major influence on gene expression, the extent to which cis-acting DNA sequences contribute to the specification of chromatin state remains incompletely understood. To address this, we have used a fission yeast sequence element (L5), known to be sufficient to nucleate heterochromatin, to establish de novo heterochromatin domains in the Schizosaccharomyces pombe genome. The resulting heterochromatin domains were queried for the presence of H3K9 di-methylation and Swi6p, both hallmarks of heterochromatin, and for levels of gene expression. We describe a major effect of genomic sequences in determining the size and extent of such de novo heterochromatin domains. Heterochromatin spreading is antagonized by the presence of genes, in a manner that can occur independent of strength of transcription. Increasing the dosage of Swi6p results in increased heterochromatin proximal to the L5 element, but does not result in an expansion of the heterochromatin domain, suggesting that in this context genomic effects are dominant over trans effects. Finally, we show that the ratio of Swi6p to H3K9 di-methylation is sequence-dependent and correlates with the extent of gene repression. Taken together, these data demonstrate that the sequence content of a genomic region plays a significant role in shaping its response to encroaching heterochromatin and suggest a role of DNA sequence in specifying chromatin state.
Author Summary
Epigenetic packaging of DNA sequence into chromatin is a major force in shaping the function of complex genomes. Different types of chromatin have distinct effects on gene expression, and thus chromatin state imparts distinct features on the associated genomic DNA. Our study focuses on the transition between two opposing chromatin states: euchromatin, which generally correlates with gene expression, and heterochromatin, which is typically refractive to gene expression. While heterochromatin is capable of spreading into euchromatic domains, the parameters that influence such spreading are unknown. We established heterochromatin at ectopic sites in the genome and evaluated whether specific DNA sequences affected the extent of heterochromatin spreading and the transition between heterochromatin and euchromatin. We found that the nature of the genomic DNA neighboring the heterochromatic sequence dramatically affected the extent of heterochromatin spreading. In particular, the presence of genes antagonized the spread of heterochromatin, whereas neutral sequence elements were incorporated into the domain. This study demonstrates that genome sequence and chromatin identity are inextricably linked; features of both interact to determine the structural and functional fate of underlying DNA sequences.
Introduction
Correct patterns of gene expression are established by orchestrated interactions among cis-regulatory elements, trans-acting factors and the surrounding chromatin environment. How these interactions are coordinated and to what extent genomic sequence serves as a blueprint, directing these interactions towards normal growth and development, remain major questions in genome biology.
Chromatin has classically been divided into two functionally distinct types: heterochromatin and euchromatin. Genes inserted within, or proximal to, major heterochromatin domains can exhibit either variegated or complete silencing [1]–[7]. This repression, referred to as position effect variegation (PEV), results from the propagation of heterochromatin marks along the chromosome, placing the euchromatic gene into a chromatin context that is incompatible with normal gene expression [2],[8],[9]. While PEV and the factors that contribute to it have been most thoroughly elucidated in yeast and flies, position-dependent gene silencing has been observed in a range of organisms including both mice and humans [1]–[3],[10]. Indeed, there are examples of human disease that can be attributed to gene silencing associated with aberrant formation of heterochromatin [11]–[14]. Together, these studies highlight the important relationship between chromatin context and gene expression and suggest that eukaryotes have developed mechanisms to counter the spread of repressive heterochromatin [2],[8],[15],[16]. However, the nature of these mechanisms and the extent to which they utilize specific DNA sequences remains incompletely understood.
Several studies have pointed towards the importance of genome sequence in shaping epigenetic states. For example, insulators are specific DNA sequences that protect genes from the regulatory effects of neighboring domains, thus enforcing domain boundaries [17]. As presently defined, insulator activity has two components: the ability to prevent cross-talk between an enhancer and promoter (enhancer blockers) and the ability to stop the spread of repressive heterochromatin (heterochromatin barriers) [17]–[20]. First identified and characterized in flies [18],[21], insulators have since been identified in vertebrates [22]–[24].
Elucidating the role of genome sequence in shaping chromatin domains requires an experimental system in which heterochromatin nucleation can be initiated in a controlled manner. To this end, we have examined heterochromatin spreading from a de novo nucleation site in the fission yeast, Schizosaccharomyces pombe. The unique advantage of this system, in addition to its genetic tractability, is the presence of well-defined DNA sequences, referred to here as heterochromatin-nucleating sequences, that are sufficient to induce heterochromatin formation de novo [25]–[27]. Moreover, introduction of a de novo heterochromatin domain at a euchromatic locus permits a simplified view of this process, in contrast to native domains of heterochromatin that result from the complex interplay of multiple sites of nucleation and heterochromatin barriers [27]–[29]. Analysis of the resulting de novo heterochromatin domains clearly implicates primary DNA sequence in defining both the magnitude and extent of the heterochromatin domain. The conceptual framework that emerges from this study provides a basis for exploring the nature of complex genomes and the impact of genome sequence on the establishment and maintenance of chromatin domains, in organisms ranging from yeast to mammals.
Results
The L5 element nucleates a de novo heterochromatin domain encompassing adjacent genomic sequences
Previous studies in S. pombe have demonstrated that a fragment of pericentromeric DNA, called L5, is capable of nucleating heterochromatin, marked by di-methylation at H3K9 (H3K9me2) and the presence of the HP1 homologue, Swi6p, at an ectopic site through an RNAi-dependent mechanism [25],[30]. Integration of the L5 element leads to the repression of an adjacent reporter gene in a manner that appears largely similar to that observed at the endogenous centromere [25],[27],[30]. What is unknown, however, is the extent to which L5-nucleated heterochromatin is capable of extending past the reporter construct into endogenous genomic sequences.
To address this question, we created a construct containing the 1.6 kb L5 element upstream of an ade6+ reporter gene. This construct was then integrated at the euchromatic ura4 + locus in order to create ura4::L5-ade6 + strains. In addition to the L5-containing construct, a control construct bearing only the ade6+ gene was also integrated at the ura4+ locus (ura4::ade6+). The effect of L5-integration on the chromatin environment of sequences within the ura4 locus was characterized by quantifying H3K9me2 and Swi6p levels throughout the region using chromatin immunoprecipitation (ChIP).
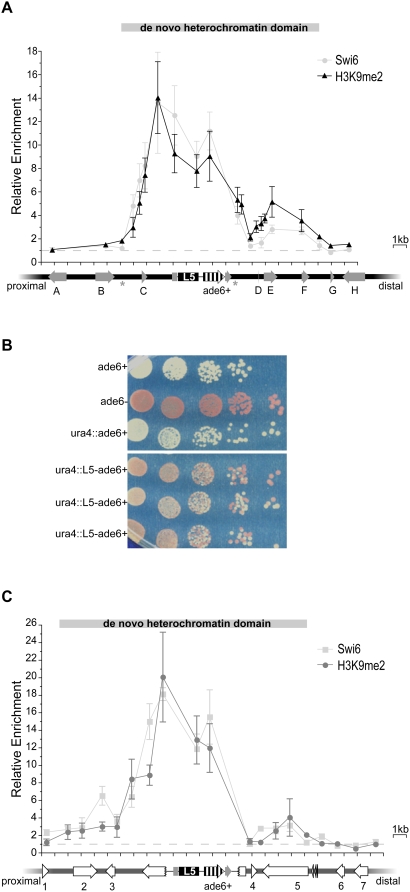
In the presence of the L5 element, H3K9me2 was enriched ∼2- to >10-fold over both the reporter gene and the surrounding genomic neighborhood (Figure 1A and Figure S1A), extending 4 kb proximal and 10 kb distal to L5. The pattern of Swi6p enrichment is remarkably similar to the level of H3K9me2, consistent with previous reports demonstrating that H3K9me2 and Swi6p have tightly overlapping distributions within heterochromatin domains (Figure 1A and Figure S1A) [28]. These data demonstrate that heterochromatin assembly is not limited to the L5-element and the reporter gene; instead, heterochromatin spreads bi-directionally into adjacent, formerly euchromatic, sequences, resulting in a de novo heterochromatin domain that spans approximately 15 kb. Throughout, we will describe the properties of a heterochromatin domain by its extent, the distance over which heterochromatin is enriched, and its magnitude, the level of heterochromatin enrichment at a given location.
Figure 1. L5 initiates formation of de novo heterochromatin domains at two distinct loci.
(A) Levels of H3K9me2 (black) and Swi6p (grey) were assayed via ChIP and are shown relative to levels at act1+, such that the dotted line indicates no enrichment. The enrichment data are plotted versus the ura4 genomic region; grey arrows represent genes, each given a letter identifier, and the direction of the arrowhead indicates the direction of transcription. Non-coding RNAs are shown as asterisks. The disrupted ura4 gene is shown as a broken arrow surrounding L5 and ade6+. Tick marks are spaced every 1 kb. The de novo heterochromatin domain is defined as regions that are greater than 2-fold enriched in H3K9me2. Error bars represent the standard error of the mean. (B) ade6+ expression is reduced in strains containing L5. Shown is the phenotypic assessment of ade6+ expression using a serial dilution assay. Each row represents an individual strain plated on adenine limiting media. ade6+ and ade6− strains demonstrate the phenotypic effects of ade6 expression and are compared to a representative ura4::ade6 + control strain and three independent ura4::L5-ade6+ strains. (C) H3K9me2 and Swi6p enrichment are plotted in dark and light grey, respectively, at the spbc2f12.03 locus. Genes in this genomic location are labeled numerically and are represented by open arrows. The disrupted spcbc2f12.03 gene is represented by a broken arrow.
The presence of heterochromatin causes reduced expression within the de novo domain
Because the chromatin state of genes near ura4 changes upon insertion of L5, we sought to determine whether gene expression at the ectopic locus was also altered. Quantitative RT- PCR (qRT–PCR) was used to quantify the levels of mRNA in the presence of L5 relative to control strains lacking L5. As expected from earlier studies [25], we observed an L5-dependent decrease in ade6+ expression; however, the reduction in expression was moderate (34±5%), indicating that silencing is incomplete in these strains (Table 1). In addition to ade6+, two genes within the de novo heterochromatin domain, located 2.7 kb proximal and 4.9 kb distal from the L5 element, also exhibited a decrease in expression in the presence of L5, 43±10% and 52±8%, respectively. Gene expression outside of the de novo heterochromatin domain was also analyzed (Figure 1A). As predicted, three genes (A, B, H) had no significant difference in transcript abundance in the presence of L5 (Table 1). The remaining gene, G, as well as gene F that lies within the de novo domain, exhibit a discordant relationship between the enrichment of heterochromatin marks and the level of gene expression. Together, these results suggest that gene-specific features may have a greater influence on the level of gene repression, as compared to the centromere, where gene repression is more complete [31].
Table 1. de novo heterochromatin domains are associated with reduced gene expression at both the ura4 and spbc2f12.03 loci.
| Gene1 | Gene name | Distance from L5 (bp)2 | Relative mRNA (+L5/−L5) | N3 | p4 |
| A | alg115 | 8393 | 1.02±0.11 | 8 | 0.853 |
| B | CC330.07 | 6245 | 1.21±0.10 | 11 | 0.101 |
| C | spcc330.06 | 2723 | 0.57±0.10 | 14 | 0.002 |
| ade6 | 340 | 0.67±0.05 | 13 | 0.000 | |
| D | tDNAgly | 4535 | |||
| E | mug135 | 4943 | 0.48±0.08 | 8 | 0.001 |
| F | CC330.03 | 7819 | 0.90±0.20 | 17 | 0.628 |
| G | CC330.19 | 9984 | 0.66±0.03 | 8 | 0.008 |
| H | rhp7 | 10902 | 1.08±0.15 | 4 | 0.731 |
| 1 | mlo36 | 10609 | 0.83±0.09 | 5 | 0.172 |
| 2 | byr2 | 8193 | 0.67±0.15 | 6 | 0.284 |
| 3 | mrpl7 | 4875 | 0.81±0.15 | 6 | 0.289 |
| ade6 | 340 | 0.48±0.10 | 5 | 0.001 | |
| 4 | rpl1701 | 4028 | 0.58±0.08 | 5 | 0.007 |
| 5 | BC2f12.05 | 9177 | 0.62±0.18 | 6 | 0.087 |
| 6 | rpl802 | 11334 | 1.03±0.08 | 5 | 0.752 |
| 7 | ceg17 | 13164 | 1.08±0.11 | 7 | 0.335 |
From Figure 1A and 1C.
Distance of the translation start site from the nearest edge of L5.
Number of independent RNA isolations included in analysis.
p-value resulting from comparison between ura4::ade6+ and ura4::L5-ade6+ strains.
Essential for viability [82].
Mutation in mlo3 results in a growth defect [83].
Essential for viability [84].
Relative gene expression in the ura4 and spbc2f12.03 loci. Steady-state mRNA for genes in the ura4 (A–H) and spbc2f12.03 (1–7) loci were assayed via quantitative RT–PCR and are reported relative to ura4::ade6+ control strains. The distance from the transcription start site to the nearest edge of L5 is indicated for each gene.
To further explore the extent of ade6+silencing, we utilized a phenotypic assay for ade6+expression. This assay allows the extent of silencing to be resolved on a sub-colony level, as opposed to the population level queried by qRT–PCR. Under conditions of limiting adenine, yeast that are mutant, or silenced [1], for ade6+ accumulate a metabolic intermediate that results in red pigmentation. In contrast, cells in which ade6+is expressed at wild type levels remain white. Results from the phenotypic assay indicate that there is significant heterogeneity among colonies in the ura4::L5-ade6 + strains (Figure 1B). Similar to classic PEV, the colony phenotypes ranged from white to red [8]. However, distinct from PEV in Drosophila, we also observed intermediate phenotypes of pink and red with white sectors, consistent with PEV as observed in yeast [1],[32],[33].
de novo heterochromatin domains are sensitive to genomic location
We next wanted to determine whether the magnitude and extent of a de novo heterochromatin domain depends upon its location in the S. pombe genome or whether domain properties are intrinsic to the L5 element itself. To explore this, we identified a second integrant of the ura4::L5-ade6+construct on chromosome 2 (spbc2f12.03::ura4::L5-ade6+). Comparison of H3K9me2 and Swi6p enrichment at L5 and ade6+ between the two sites of integration reveals similar patterns of enrichment, suggesting that the nucleation of heterochromatin and local spreading are not sensitive to the changes in genomic location from ura4 to spbc2f12.03 (Figure 1C).
We next compared the magnitude and extent of the de novo heterochromatin domains formed at these two genomic locations. Distal to L5, the patterns of heterochromatin enrichment are markedly similar between the ura4 and spbc2f12.03 loci. In contrast, heterochromatin is observed 9 kb proximal to L5 at the spbc2f12.03 locus as compared to only 4.9 kb at the ura4 locus (compare Figure 1A and 1C). While this expansion at the spbc2f12.03 locus is only modestly enriched in H3K9me2, it is also marked by the presence of Swi6p (Figure 1C), suggesting there is a 4.1 kb expansion of the heterochromatin domain relative to the ura4 locus. Thus, the proximal boundary of the de novo heterochromatin domain at the ura4 locus is influenced by genomic location as opposed to reflecting an intrinsic limitation of the L5-element.
To determine whether the de novo heterochromatin domain at spbc2f12.03 alters gene expression, qRT–PCR was used to analyze mRNA levels at the spbc2f12.03 ectopic locus. Repression is observed at ade6+ and the nearby rpl1701+ gene, but not genes 2, 3 and 5 (Figure 1C, gene 4; Table 1). Thus, analogous to gene F at the ura4 locus, the recruitment of heterochromatic marks to an ectopic locus is not always associated with significant gene silencing.
Cis-acting sequences shape the de novo heterochromatin domain
The experiments described above demonstrate that the extent of an L5-dependent de novo heterochromatin domain can vary between different locations in the genome. To explore whether these differences are attributable to cis-acting factors, we engineered constructs in which different DNA sequences were placed adjacent to L5. These constructs were then inserted at the ura4 locus to examine the role of sequence in defining the heterochromatin domain, without altering its location in the genome.
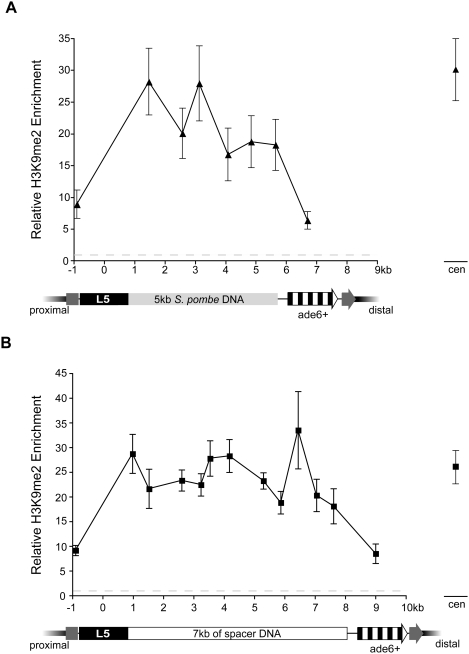
First, 5 kb of S. pombe DNA taken from a region between two divergently transcribed genes (spcc320.02+ and spcc320.03+) was positioned between L5 and ade6+. This region was selected because it is one of the larger regions in the S. pombe genome in which known protein-coding genes are absent and because it normally lacks heterochromatic modifications [28]. Having established that this region maintains the absence of H3K9me2 when moved to the ura4 locus in strains lacking L5 (Figure S2A), H3K9me2 levels were queried over this DNA in the presence of the L5 element. Heterochromatin was robustly enriched over the 5 kb insert DNA (Figure 2A). Strikingly, the magnitude of H3K9me2 enrichment is comparable to the level observed at S. pombe centromeres (Figure 2A), suggesting that H3K9me2 can reach and sustain high levels of occupancy over the entire region. This is in contrast to the pattern of heterochromatin spreading over the gene-rich ura4 and spbc2f12.03 neighborhoods (Figure 1A and 1C). The difference between the magnitudes of heterochromatin enrichment between these DNA sequences supports the role of cis-acting DNA sequences, potentially the genes themselves, in shaping the characteristics of heterochromatin domains.
Figure 2. H3K9me2 is highly enriched over intergenic and spacer DNA fragments.
H3K9me2 enrichment is plotted versus S. pombe intergenic DNA as shown in light grey (A) and DNA from the phage lambda as shown in white (B). The dark grey arrow represents the disrupted ura4 gene. The data points on the far right represent the level of H3K9me2 enrichment observed at the pericentromeric repeats in the respective strains, as internal controls.
Consistent with this hypothesis, there is a significant reduction in H3K9me2 enrichment coincident with the start of the ade6+ gene, and the level of enrichment at this location is similar to the level observed when ade6+ is adjacent to L5 (compare Figure 1A and 2A). One interpretation of these data is that only low levels of heterochromatin can exist in transcriptionally active regions. Thus, when heterochromatin spreads from the spacer DNA into the ade6+ gene, the ade6+ gene behaves as a heterochromatin attenuator. Alternatively, this may indicate that the extent of spreading is constrained such that heterochromatin cannot spread, with high levels of enrichment, farther than 5.6 kb from L5.
To address this latter possibility, a longer spacer sequence was selected and inserted between L5 and ade6+. Sequences from lambda phage were chosen, as they have been used in previous epigenetic studies as spacer DNA [34]. No significant enrichment was observed over the length of the 7 kb insert in the absence of L5, suggesting that these sequences do not nucleate heterochromatin on their own (Figure S2B). In contrast, when the L5 element is present, robust enrichment in H3K9me2 was observed over the length of the lambda fragment, at levels similar to that of the centromeres and the 5 kb S. pombe spacer fragment (Figure 2B). Moreover, when we extended our analysis to include the levels of H3K9me2 enrichment at endogenous sequences in ura4::L5-7kb-ade6+ strains we found that it was remarkably similar to the levels observed in strains lacking spacer DNA (Figure S3). Thus, the addition of spacer DNA (up to 7 kb) does not constrain the extent of a de novo heterochromatin domain. Instead, our data are consistent with a model in which both the extent and magnitude of a heterochromatin domain are dictated by features of the underlying DNA sequence.
The boundaries of de novo heterochromatin domains are marked by the presence of highly transcribed genes
Because endogenous sequences can influence the extent of a heterochromatin domain, we next sought to determine the factors that mediate the interaction of DNA sequence and chromatin. Based on the observation that some barriers require formation of a transcription complex [22],[27],[29],[35],[36], we investigated the relationship between domain size and transcriptional activity.
The level of transcriptional activity within both the ura4 and the spbc2f12.03 regions could be assessed using previously reported data sets [29],[37]. Transcriptional activity was inferred from both the steady state levels of mRNA and the level of RNA Polymerase II (Pol II) and RNA Polymerase III (Pol III) enrichment at the promoter. Between the two regions, there were five loci that were transcriptionally exceptional: one gene that was transcribed by Pol III and four genes with unusually high levels of Pol II transcriptional activity (Table 2) [29],[37].
Table 2. Genes within the de novo heterochromatin domains are highly expressed and enriched in Pol II.
| Gene | Steady state mRNA levels1 | RNA Pol II Enrichment1 |
| A | 1769 | 1.14 |
| B | 761 | 0.91 |
| C | 10588 | 3.41 |
| E | 618 | 0.76 |
| F | 377 | |
| G | 296 | 0.64 |
| H | 942 | 0.88 |
| 1 | 8746 | 3.28 |
| 2 | 347 | 0.64 |
| 3 | 1885 | 0.88 |
| 1845 | 1.47 | |
| 4 | 9130 | 3.81 |
| 5 | 2220 | 0.88 |
| 6 | 9810 | 4.75 |
| 7 | 364 | 0.80 |
| ade6+ | 5472 | 0.91 |
From [37].
Transcription levels at the wild type ura4+ and spbc2f12.03+ loci.
The ura4 genomic neighborhood includes a Pol III-transcribed tDNAGly (gene D in Figure 1A), which is coincident with an H3K9me2 gap. This gap could be attributed to general nucleosome depletion or, alternatively, to nucleosomes shielded from H3K9me2 modification by the Pol III transcription complex. Supporting the former hypothesis, tDNA genes are generally depleted of nucleosomes when compared to the genome average [38]. To distinguish between these two possibilities, an antibody to the C-terminus of histone H3 was used to characterize nucleosome occupancy surrounding to the tDNAGly gene. Indeed, the level of H3 enrichment at the tDNAGly was reduced relative to sequences in the surrounding neighborhood (Figure S1B), indicating that the observed H3K9me2 gap is due to decreased nucleosome occupancy surrounding the tDNAGly.
The ura4 genomic neighborhood includes the gene spcc330.06+(gene C in Figure 1A), which is highly expressed and enriched in Pol II (at the 94th percentile genome-wide) at its promoter (Table 2) [37]. This gene is located within a striking transition in H3K9me2 enrichment from 14-fold to <2-fold enrichment over a distance of only 2.7 kb (Figure 1A). In contrast to the nucleosome gap discussed above, this transition marks a boundary of heterochromatin enrichment and cannot be explained by reduced nucleosome occupancy (Figure S1B). We hypothesize that this gene may behave as a heterochromatin barrier and, more broadly, that highly expressed genes in general may be effective heterochromatin barriers.
Within the spbc2f12.03 genomic neighborhood, three genes are highly transcribed (genes 1,4 and 6 in Figure 1C). One of these genes (gene 6) is distal to the boundary of the de novo heterochromatin domain, and as such is uninformative. However, genes 1 and 4 (Figure 1B) are located at boundaries of the de novo heterochromatin domain, consistent with the hypothesis that highly expressed genes weaken and/or stop the spread of de novo heterochromatin.
Introduction of a gene within spacer DNA attenuates heterochromatin spreading independent of level of transcription
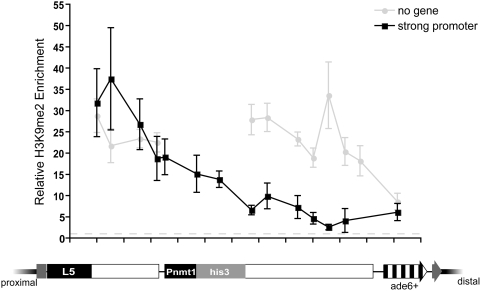
To directly test whether the presence of transcribed genes can influence the extent of a de novo heterochromatin domain, we constructed a chimeric reporter gene composed of the strong, repressible, nmt1+ promoter driving expression of the his3+ open reading frame (Pnmt1-his3+) [39]. This construct was then inserted within the 7 kb spacer fragment, and heterochromatin spreading was monitored over the spacer sequences and the inserted gene. As expected, H3K9me2 was highly enriched over the spacer DNA proximal to Pnmt1-his3+, consistent with the levels observed in uninterrupted spacer strains (Figure 3). However, the magnitude of H3K9me2 enrichment decreases over the Pnmt1-his3+sequences and remains reduced over the distal portion of the spacer DNA (Figure 3). These data demonstrate that the insertion of genic sequences within the spacer DNA attenuates the spread of heterochromatin and further support the hypothesis that the presence of genes within the ura4 and spbc2f12.03 neighborhoods limits heterochromatin spreading. It is interesting, however, that the Pnmt1-his3+ construct, despite being more highly transcribed than spcc330.06+ (Figure S5A), does not exhibit complete barrier activity (Figure 3), suggesting that factors other than high levels of transcriptional activity are required for complete barrier activity.
Figure 3. The presence of Pnmt1-his3 attenuates heterochromatin spreading.
H3K9me2 enrichment in is shown for strains in which Pnmt1-his3+ has been inserted within lambda (black) or containing uninterrupted spacer DNA (grey).
Because the presence of genes antagonizes heterochromatin spreading, we sought to determine whether a high level of transcriptional activity is required for attenuator activity. To test this we took advantage of an engineered allele of the nmt1+ promoter that results in reduced transcription efficiency [40], and cultured these strains in medium containing thiamine, which results in further repression of the nmt1+ promoter [39]. Despite a ∼570 fold decrease in expression the weakened Pnmt1-his3+ gene still exhibited significant attenuation ability, indistinguishable from the strongest allele (Figure S5B). Thus, other features of the nmt1+ promoter may serve to attenuate the spread of heterochromatin. Indeed, the region of the promoter that is required for thiamine repression binds a protein complex independent of thiamine conditions [41]. This protein complex, or other complexes that localize to the promoter independent of thiamine and transcription efficiency, may serve to attenuate the spread of heterochromatin.
Increased L5-copy number does not alter the heterochromatin domain
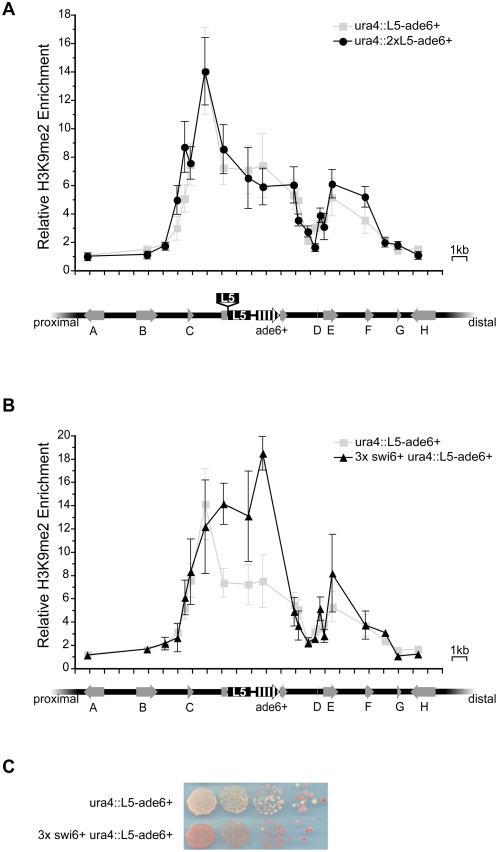
Having demonstrated the impact of genome sequence on the extent of spreading from a heterochromatin-nucleating sequence, we wanted to determine whether changes to the sequence content, in terms of L5 copy number, would alter the properties of a de novo heterochromatin domain. Thus, two copies of L5 were inserted in tandem at the ura4 locus. The magnitude and extent of heterochromatin enrichment in these strains was markedly similar to strains bearing one copy of L5 (Figure 4A), suggesting that the copy number of L5 does not notably enhance heterochromatin enrichment or spreading within a de novo domain.
Figure 4. The de novo heterochromatin domain is shaped by the dosage of trans-acting factors.
(A) H3K9me2 enrichment in the presence of an additional copy of L5 is shown in black circles. The extra copy of L5 is shown above the ura4 genomic region, with insertion site indicated, compared to strains bearing only one copy of L5 (light grey squares). (B) H3K9me2 enrichment in strains bearing extra copies of the swi6+ gene, shown in black triangles. (C) Serial dilution analysis of ade6+ expression in wild type and 3× swi6+ strains.
The boundaries of the de novo heterochromatin domain are insensitive to increased dosage of swi6+
We also wanted to address the possible role of trans-acting factors in regulating the extent of the heterochromatin domain, either by competition with other heterochromatic regions for limiting heterochromatin components [42] or by competition between heterochromatic and euchromatic factors for the same nucleosome substrate [43]. We hypothesized that increasing the dosage of heterochromatin proteins (or reducing the amount of competing factors) should result in the expansion of a heterochromatin domain [9], [42]–[45].
Swi6p is a dosage-dependent modifier of heterochromatin levels at the S. pombe mating-type locus as well as a limiting factor in heterochromatin formation [46],[47]. Thus, we analyzed the magnitude and extent of the ura4 de novo heterochromatin domain in strains bearing three copies of swi6+ [47]. We confirmed that the level of swi6+ mRNA is increased by 2.7-fold in these strains (data not shown). While the local magnitude of H3K9me2 proximal to L5 was increased in these strains (Figure 4B), the increased dosage of swi6+ did not result in the expansion of the heterochromatin domain. Consistent with the increased level of H3K9me2 enrichment, ade6+ expression was further reduced in these strains, resulting in an increased proportion of red colonies (Figure 4C and Figure S4). In contrast, increased swi6+ dosage did not significantly affect expression of other genes within the ura4+ neighborhood (Figure S4). This suggests that, while the level of Swi6p influences both the local concentration of H3K9me2 and the level of gene expression, the extent of the de novo heterochromatin domain is not sensitive to increased dosage of swi6+.
Functional distinction between local heterochromatin formation and spreading over spacer DNA
In the absence of known transcribed elements, H3K9me2 spreads unattenuated over distances at least up to 7 kb (Figure 2B), resulting in a consistent level of H3K9me2-enrichment at ade6+ independent of the presence of spacer DNA (Figure 5C). What remains to be addressed is whether the level and the stability of gene silencing differ between strains containing spacer DNA versus ura4::L5-ade6+.
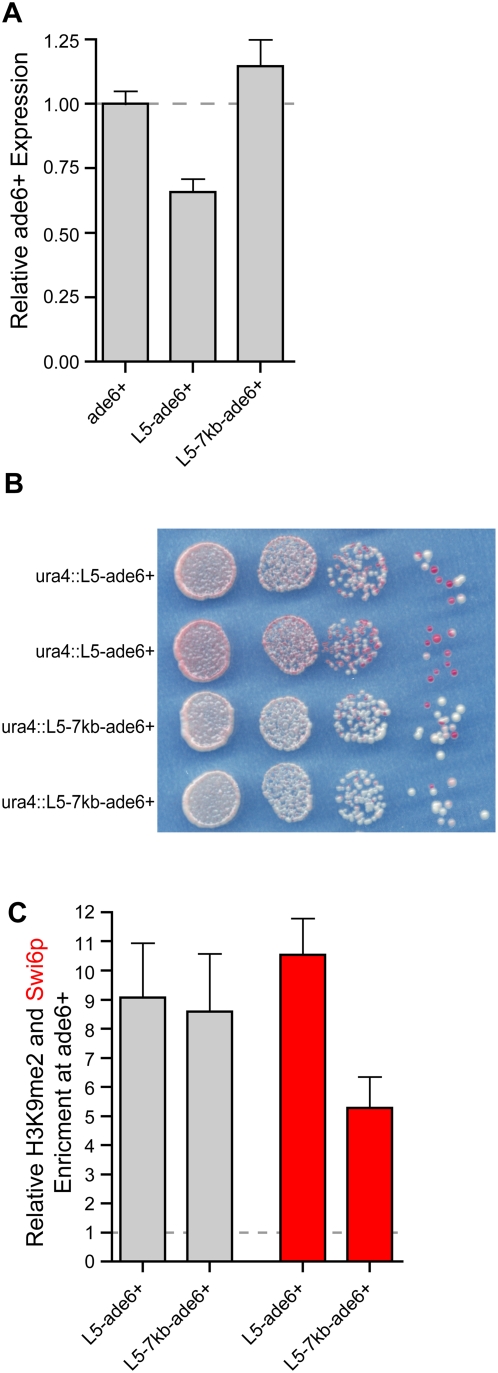
Figure 5. Local versus spreading over spacer DNA exerts different effects on ade6+ expression.
(A) Levels of ade6+ expression in control (ura4::ade6+) strains compared to both ura4::L5-ade6+ and ura4::L5-7kb-ade6+strains. (B) Serial dilution assay comparing the extent of ade6+ expression in both ura4::L5-ade6+ and ura4::L5-7kb-ade6+strains. (C) The level of H3K9me2 enrichment at ade6+ in ura4::L5-ade6+ and ura4::L5-7kb-ade6+ strains is shown as gray bars. The level of Swi6p enrichment is plotted in red.
To explore this question, we compared the levels of ade6+expression by qRT–PCR and found that when ade6+was located 7 kb away from the L5 element, distal to the lambda spacer, silencing was no longer observed despite the presence of H3K9me2 (Figure 5A and 5C). This finding was confirmed using the phenotypic ade6+ assay, which revealed a much lower level of silencing in ura4::L5-7kb-ade6+ strains (Figure 5B). These data suggest that, even when the levels of H3K9me2-enrichment are similar (Figure 5C), heterochromatin formed proximal to L5 and heterochromatin formed over spacer DNA can have different effects on gene expression.
We suspected that the differences in silencing could be attributed to the levels of Swi6p at the ade6+ gene in ura4::L5-ade6+ as compared to ura4::L5-7kb-ade6+ strains. To address this hypothesis, we assessed the level of Swi6p enrichment over the lambda spacer DNA and distal ade6+ gene. We observed a significant decrease in Swi6p enrichment relative to the level of H3K9me2 across spacer and ade6+ DNA when compared to ura4::L5-ade6+ and spbc2f12.03::ura4::L5-ade6+ strains, as well as to other heterochromatic loci (Figure S6 and Figure 5C). This reduction is consistent with the decreased levels of silencing and could be a function of long distance spreading or a sequence-dependent affect of spacer DNA.
In addition to the total level of gene silencing, another manner in which the reduced levels of Swi6p in spacer strains could affect gene expression is by altering the stability of gene repression. When transgenes are placed within the centromere, or at locations throughout the mating type locus, their phenotypic stability (silenced or expressed) can vary with location and Swi6p dosage [31],[32],[46],[47]. To address whether the silenced and expressed states are stable to equivalent degrees in cases of local (high levels of Swi6p) versus spreading over spacer DNA (reduced levels of Swi6p) we chose colonies that were either silenced or expressed, as determined by their ade6+ expression phenotype (ie. entirely red or entirely white, respectively). The stability of the silenced state was determined by the proportion of progeny that exhibited silencing after a period of overnight growth. We examined the phenotypic stability of ade6+ and ade6− controls, and as expected, the progeny maintained the appropriate phenotype (Figure 6A). However, when expressed colonies were chosen from ura4::L5-ade6+ strains, only ∼62% maintained the completely expressed phenotype, while the remaining colonies switched to a partially or completely silenced phenotype. This is in stark contrast to the ura4::L5-7kb-ade6+ strains, in which 95% of the progeny maintained the expressed state (Figure 6B), suggesting that the establishment of silencing (i.e., switching from an ade6+ expressed state to a silenced state) occurs less often when ade6+ is distal to 7 kb of spacer DNA and less enriched in Swi6p, despite comparable levels of the epigenetic mark H3K9me2.
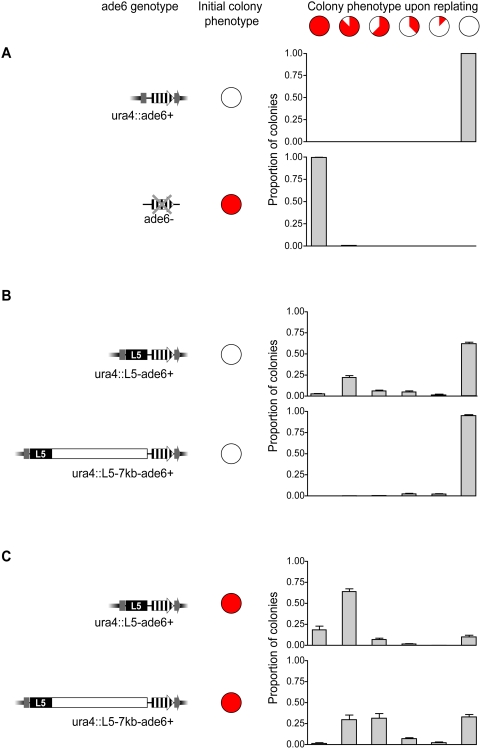
Figure 6. The extent and stability of ade6+ silencing is altered in spacer strains.
(A–C) Colonies were selected on the basis of the initial colony phenotype (either all entirely red or entirely white) for each of the given genotypes. After a 24-hour period of growth, strains were re-plated and the phenotypes of the resultant colonies were scored and classified based on the proportion of the colony that exhibited silencing, as indicated schematically above the graphs. Intermediate silencing phenotypes included sectoring and homogenous, intermediate levels of pigmentation. The graphs below represent the proportion of colonies in each class based on genotype and initial colony color.
When silenced colonies were selected from ura4::L5-ade6+ strains, ∼18% of the progeny exhibited phenotypes indicative of complete silencing and 90% exhibited at least partial silencing. In contrast, silenced colonies from ura4::L5-7kb-ade6+ strains were less likely to give rise to progeny that exhibited complete or partial silencing (∼1% and 69%, respectively), suggesting that maintenance of silencing is also less frequent when ade6+ is separated from L5 by spacer DNA and reduced in Swi6p localization (Figure 6C). These data provide evidence that the level of Swi6 impacts the establishment and maintenance of silencing, despite consistent levels of H3K9me2.
Discussion
Ectopic gene silencing and/or heterochromatin formation has previously been studied in mammalian systems [48]–[51]. Ectopic X inactivation, for example, has been shown to affect gene expression on a large scale [52]. Typically, however, the complex nature of the mammalian genome restricts the focus of these studies to local heterochromatin formation and single gene repression. In this study, the compact nature of the S. pombe genome and our ability to robustly query for the presence of heterochromatin allowed us to rigorously test the response of multiple DNA sequences to encroaching heterochromatin. Our data demonstrate a clear effect of genomic sequence in shaping both the extent and magnitude of a heterochromatin domain and demonstrate that, while the eukaryotic genome is permissive to the negative transcriptional effects of heterochromatin, euchromatic sequences can counteract encroaching heterochromatin.
de novo heterochromatin domains are shaped by DNA sequences that vary in their ability to promote or antagonize heterochromatin spreading
The relationship between the size of a heterochromatin domain and the presence of specific heterochromatin barriers has been previously established in a number of eukaryotic organisms [53]. Our study extends this conclusion, demonstrating that DNA sequences exert a range of effects on heterochromatin domains. For example, the ade6+ gene dampens heterochromatin enrichment independent of both genomic location and distance from L5, but is insufficient to completely stop heterochromatin spreading (Figures 1A, 1C, 2A, and 2B). In contrast, intergenic and spacer DNA sequences promote the assembly of robust H3K9me2. We propose that there is a spectrum of effects, ranging from antagonistic to cooperative, that genomic sequence can exert on heterochromatin (Figure 7). This model incorporates the complexity and context dependence of genomic sequence and its relationship to heterochromatin and is applicable to sequences in yeast, as seen here, or in more complex genomes, as will be discussed below.
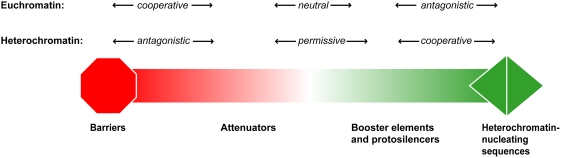
Figure 7. The continuum of DNA sequence, its effects on heterochromatin spreading, and the balance between opposing heterochromatic and euchromatic forces.
The range of DNA sequences and the magnitude of their interaction with DNA is depicted along a gradient ranging from heterochromatin barriers (red) to heterochromatin nucleating sequences (green). The italic text above the figure describes the effect of a specific DNA sequence on both heterochromatin and euchromatin. Below the figure is a classification system that subdivides the continuum.
For this model, we have subdivided the discrete extremes of DNA sequences noted previously (that is, heterochromatin-nucleating sequences and heterochromatin barriers) into subclasses that include attenuators, neutral elements and protosilencers/boosters. While this is helpful for purposes of discussion, we do not wish to impose strict definitions, especially in light of data from this study, suggesting that particular sequences may be placed at multiple points along the continuum, depending on their context.
Heterochromatin antagonists: barriers and attenuators
Our data confirm that heterochromatin can spread from L5 in both directions over euchromatic DNA, resulting in a de novo heterochromatin domain encompassing multiple endogenous genes and altering gene expression (Figure 1A and 1C, Table 1). However, gene repression within the de novo domain is moderate, at most about 50%. The incomplete silencing observed within de novo heterochromatin domains, as well as the boundaries of these domains, may be a consequence of the factors present within euchromatic domains that antagonize the propagation of heterochromatin.
The boundaries of de novo heterochromatin domains are marked by three highly transcribed genes, implicating Pol II transcription in barrier activity (Figure 1A and 1C, Table 2) [37]. The ade6+ gene is also transcribed and enriched in Pol II, albeit to a lesser extent than the three putative barriers. These four sequences may rely on transcription to counteract the spread of heterochromatin from the L5 heterochromatin-nucleating sequences. However, high levels of transcription are insufficient for complete barrier activity (Figure 3). Furthermore, we find that, in the case of Pnmt1-his3+, the presence of genes can attenuate the spread of heterochromatin independent of the level of transcription (Figure S5B). We conclude that DNA sequences modify heterochromatin spreading through the sequence-dependent recruitment of other mediating factors, such as transcription complexes, and dictate whether a sequence behaves as a true barrier or falls in the range of heterochromatin attenuators (Figure 7). These findings are consistent with previous results implicating transcription factors and promoters with barrier activity [35], [54]–[58]. Sequence could also influence heterochromatin directly, as is the case with some examples of nucleosome positioning [59] or could reflect selective pressure to maintain domain boundaries (Table 1).
In addition to protein-coding genes, the ura4 de novo heterochromatin domain includes a tDNAGly gene (discussed below) and non-coding RNAs (Figure 1A). The ura4 locus is not unique in its transcriptional makeup, as recent studies have provided insight into the vast amount of transcription occurring in the S. pombe genome outside of canonical protein coding genes [60],[61]. Additionally, the ura4 neighborhood also includes solo long terminal repeats (LTRs) [62]. How these features interact with heterochromatin spreading, and whether they shape the formation of de novo heterochromatin domains warrants further genome-wide studies.
Transcription by Pol III complexes has an established relationship with barrier activity in yeast genomes [27],[29],[35],[36]. A tDNAAla within the S. pombe centromere 1 prevents the spread of heterochromatin into the abutting domain of centromeric chromatin. In contrast, the tDNAGly gene is not coincident with the domain boundary of the ectopic heterochromatin domain formed at ura4+; however it is deficient in H3K9me2 enrichment, due to the absence of a nucleosome(s) (Figure 1A and Figure S1B). Nucleosome depletion has been shown previously to restrict heterochromatin spreading [63]; in this context, the nucleosome gap may weaken the spread of heterochromatin, resulting in the gradual attenuation observed distal to the tDNAGly. We suggest that, like the ade6+ gene, the tDNAGly behaves as heterochromatin attenuator in our experimental system. It is interesting to note that, while other tDNAs substituted at the centromere recapitulate barrier activity, re-positioning of tDNAAla at a euchromatic locus resulted in an attenuation of heterochromatin spreading, but not complete barrier activity [27]. Together, these data establish a mechanistic link between heterochromatin barriers and attenuators, and implicate genomic context as an additional factor in determining where a sequence falls along the continuum of effects on heterochromatin (Figure 7).
DNA sequences prevent expansion of heterochromatin domains
Whether the effect of DNA sequence could be abrogated by increased dosage of heterochromatin proteins was also examined. Increased swi6+ resulted in increased levels of H3K9me2 over sequences adjacent to L5, as well as enhanced repression of ade6+, consistent with an increase in local heterochromatin (Figure 4B). However, this change in heterochromatin enrichment is not accompanied by an expansion of the domain. Further expansion of the domain is likely prevented by the barrier and attenuator activity of adjacent sequences, indicating that these sequences are robust to the increasing magnitude of heterochromatin in these strains. This is the also the case with models of PEV in mouse where enhancing heterochromatin formation is insufficient to cause PEV when a transgene is flanked by chromatin insulators [64]. Alternatively, enhanced propagation of heterochromatin could be limited by selection against increased silencing of genes within the de novo heterochroamtin domain.
Active and passive heterochromatin advocates: protosilencers, boosters, and neutral sequences
As heterochromatin antagonists are characterized by different strengths, we propose that DNA sequences also differ in their ability to initiate or promote heterochromatin spreading. The identification of protosilencers, sequences that can actively contribute to gene silencing but only in specific “silencing-conducive” environments [65], supports this hypothesis. DNA sequences that are permissive to heterochromatin spreading can be conceptually subdivided into those that rely on active mechanisms, like those above, and those that passively allow heterochromatin but do not actively propagate the heterochromatic state (Figure 7). The spacer and S. pombe intergenic fragments may fall into this class of sequence elements. Both sequences allow formation of large heterochromatin domains with levels of H3K9me2 enrichment similar to that observed at the centromere (Figure 2A and 2B). Alternatively, these sequences may contain elements that enhance heterochromatin spreading, and thus would belong in the former class of sequences that actively promote heterochromatin spreading.
Interestingly, while high levels of H3K9me2 are sustained over the length of the lambda spacer DNA, the ratio of Swi6p/H3K9me2 is reduced, relative to both genome-wide data and data from the ura4 and spbc2f12.03 de novo heterochromatin domains (Figure 5C and Figure S6) [28]. The reduced levels of Swi6p correlate with reduced ability to establish and maintain silencing at ade6+ when compared to ura4::L5-ade6+ strains (Figure 6). These data suggest that lambda spacer DNA exerts a sequence-specific effect on the associated heterochromatin domain that results in reduced levels of gene repression.
Genome sequence affects chromatin state in higher eukaryotes, as well as fission yeast
The spreading of heterochromatin from L5 shares at least conceptual similarities with the spreading of gene silencing and, presumably, facultative heterochromatin from an ectopic X inactivation center in mammalian systems [52],[66]. Furthemore, as we demonstrate in fission yeast, genome sequence is also implicated in the organization of chromatin on the mammalian X chromosome [67]. The inactive X chromosome is organized in alternating domains of genes that are subject to inactivation (silenced) and domains of genes that escape from X inactivation (expressed) [68],[69], as well as by domains of different types of heterochromatin [70],[71]. A CTCF site on the mouse inactive X chromosome, located within such a transition region, exhibits insulator activity in transgene assays [23], thus implicating DNA sequence in maintaining the boundaries of expression domains. Moreover, the presence of specific DNA features on the X chromosome can be used to accurately predict whether a gene will be subject to, or escape from, X inactivation [72],[73]. However, as with the intergenic and spacer fragments in this study, it is unknown whether the sequences correlated with gene silencing passively permit the silent state, or whether they actively promote the propagation of gene silencing. Finally, LINE-1 elements have been proposed to behave as protosilencers, or booster elements, relaying transcriptional inactivation from sites of nucleation [72],[74]. While such evidence points to the importance of DNA sequence in regulating domains of gene expression on the X chromosome, the presence of barriers and other sequences in mammals has yet to be addressed fully.
Materials and Methods
Fission yeast strains
The genotypes for strains used in this study are as listed (Table S1). Fission yeast media were prepared using standard procedures [75]. For repression of the nmt1 promoter 15 uM thiaimine was added [76]. A strain bearing the ade6DN/N allele (a loss of function mutation created by a 153 bp deletion of the ade6+ open reading frame [77]) was generated (Kfy539) and was transformed via electroporation (1.5 kV, 200Ω, 25uF) on a BioRad Gene Pulser II. Transformed cells were selected on PMG media lacking adenine [75]. Colonies derived from strain Kfy539 were then patched onto media containing 2 g/L of 5-fluoro-orotic acid (FOA) (MP Biomedicals) to select for disruption of ura4+. The resulting strains were screened, using Southern analysis, for appropriate integration of ade6+. Additionally, BW17 transformants were screened by Southern blot for the maintenance of the 7 kb lambda DNA fragment. At least three independent transformants of each genotype were maintained (with the exception of the random integrant, Kfy812) and used for further analysis. All transformants were then crossed into a swi6+ strain and the ura4::L5-ade6+ allele was selected for on the basis of FOA resistance. To create swi6+333 strains, ura4::L5-ade6+ strains were crossed into SPG1232 (Shiv Grewal) [47]. To create ura4::L5-7kb::(Pnmt1-his3+)-ade6+ the Pnmt1-his3+ construct was transformed into Kfy589, colonies were selected for on the basis of growth on media lacking histidine. After integration within lambda was confirmed by Southern analysis, these strains were crossed into a swi6+ strain.
Plasmids
To create plasmid BW5, ade6 + was amplified from S. pombe genomic DNA using primers BWP34F and BWP34Rb (Table S2) to add StuI, SpeI, ClaI, and BglII sites to the 5′ end of the product and Sac1, Sma1 and Stu1 sites to the 3′ end. The PCR product was then digested with StuI and inserted into the StuI site of ura4+ in pUC13/18. The ade6+ open reading frame and upstream region were sequenced to ensure no mutations had been introduced during cloning.
Plasmid BW7 was constructed through digestion of YL317 with SpeI and ClaI and subsequent purification of the L5-containing fragment [27]. L5 was then inserted into the SpeI/ClaI site of BW5. Plasmids BW32 and BW34 contain 4.9 kb of S. pombe intergenic DNA taken from between SPCC320.02 and SPCC320.03 inserted into the BglII site of BW5 and BW7, respectively. The intergenic fragment was digested from the cosmid SPCC320 using XbaI, subcloned into pUC13/18, and then digested with BamHI before inserting into the appropriate plasmid. Plasmids BW30 and BW17 were created by digesting the lambda phage genome (NEB) with BamHI and purifying the 7.2 kb fragment, which was then ligated into the BglII sites of BW5 and BW7, respectively. To create BW20 an additional copy of L5, as a BamHI–BglII fragment, was inserted into the BglII site of BW7.
Plasmids BWP40 and BWP41 were created by replacing the GFP ORF with his3+ within the plasmids pFA6a-kanMX6-P3nmt1-GFP and pFA6a-kanMX6-P41nmt1-GFP, respectively (A gift from Jian-Qiu Wu) [78]. A subfragment of the lambda spacer DNA was liberated from BW17 by digestion with BglII and cloned into pUC1318. The Pnmt1-his3+ containing fragment was then inserted within the PstI site in the lambda fragment.
Confirming and mapping random integrants
To identify random integrants that did not disrupt the ura4 locus, we selected transformants on the basis of growth on PMG–adenine and death on FOA. These strains were then confirmed via Southern blot to have a single ade6+ insertion and the site of integration was mapped using an inverse PCR protocol modified from [79]. Genomic DNA (2 µL) was digested with MboI or Nde1 and incubated for 3.5 hours at 37°C. The digest was heat inactivated at 65°C for 20 minutes. 2 µL of the digest was added to a standard ligation reaction (T4 ligase, NEB) and incubated overnight at room temperature. Inverse PCR was performed using primers E367/BWP89F for the Nde1 digest and BWP37F/BWP32F for the Mbo1 digest. The PCR products were purified and sequenced using the PCR primers listed above.
Serial dilution analysis and scoring of ade6+ phenotypes
Strains were grown overnight with shaking in YES media at 32°C and diluted to a concentration of 1e6 cells/mL. Cultures were diluted serially (1∶9) and plated on YES media lacking adenine.
To assess the stability of silencing, colonies that were scored as either completely white or completely red were identified using a Leica MZ7.5 microscope and grown for 24 hours in YES media before plating on YE plates lacking adenine.
For both protocols, plates were grown for three nights at 32°C and shifted to 4°C for 24 hours before photographing or counting.
Real time RT–PCR
Total nucleic acid was isolated from logarithmically growing cells in YES media at 32°C, and was then subjected to DNAse treatment and RT–PCR using oligodT as a primer. Expression was analyzed by quantitative PCR using SYBR Green on a Bio-rad myCycler, using primers specific to the wild type copy of ade6+ (BWP85F/R). Levels of mRNA from ade6+, and other genes queried, were expressed relative to act1+(BWP74F/R). The standard curve was generated using genomic DNA isolated from strain Kfy1. In order to be included in this study a PCR experiment had to have a PCR efficiency between 90–110% and a correlation coefficient >0.99.
Chromatin immunoprecipitation
The H3K9me2 ChIP protocol was adapted from [80]. Logarithmically growing cells from control and experimental strains were treated with 1% paraformaldehyde for 15 minutes. The cell wall was then destroyed through bead beating twice for two minutes in buffer containing protease inhibitors. The resulting material was then sheared to an average fragment size of 600 bp using sonication. Chromatin preps were then subdivided into three tubes: an input sample that was used to check shearing, an IP sample to which protein beads and antibodies to H3K9me2 (from Takeshi Urano) were added, and a mock sample to which only protein beads were added. The mock and IP samples were incubated overnight, and the beads were isolated and subjected to a series of washes. Finally, DNA was purified from all three samples (IP, mock, and input) with phenol-chloroform extraction and ethanol precipitation using glycogen as a carrier.
H3 ChIPs were preformed as above using an antibody to H3 (abcam 1791).
Swi6p ChIPs were performed using the above protocol modified from [81]. 2.5e8 cells were shifted to room temperature for two hours prior to fixation. Cells were fixed with 3% paraformaldehyde for 30 minutes at room temperature. 1 µL of antibody (from Shiv Grewal) was incubated with the IP sample overnight, prior to incubation with protein beads for two hours.
Quantitative PCR was used to assay levels of query/act1+ in IP reactions relative to a no-antibody control.
Supporting Information
The ura4 genomic region is not enriched in H3K9me2 in the absence of L5, and exhibits varible nucleosome occupancy. (A) The ura4 locus is depicted, and assayed for the presence of H3K9me2 (black) and Swi6p (grey), in the absence of the L5 element. (B) Nucleosome occupancy was characterized using an antibody to the c-terminus of histone H3. The data are expressed relative to the nucleosome occupancy at the act1+ locus.
(3.00 MB TIF)
S. pombe intergenic and lambda spacer fragments do not recruit H3K9me2 in the absence of L5. H3K9me2 enrichment over S. pombe intergenic (A) and lambda (B) sequences in the absence of L5. The intergenic spacer DNA is in duplicate copies in the genome (at the ura4 locus as well as its endogenous locus) thus no enrichment is represented by a value of 2.
(1.88 MB TIF)
Heterochromatin spreading in the ura4 locus is unaffected by the presence of lambda spacer DNA. Analysis of H3K9me2 in the ura4 locus in ura4::L5-7kb-ade6+ strains (black) and ura4::L5-ade6+ strains. To facilitate comparison between the two strains H3K9me2 enrichment for the ura4::L5-ade6+ is depicted with a gap over the lambda insert.
(1.93 MB TIF)
Increased swi6+ copy number results in decreased ade6+ expression but does not alter expression of other genes within the de novo heterochromatin domain. Steady state mRNA levels are depicted relative to ura4::ade6+ strains for wild type strains (grey) and strains bearing 3 copies of swi6+ (black).
(1.59 MB TIF)
Insertion of a gene within lambda attenuates heterochromatin independent of the level of transcription. (A) Levels of steady state mRNA relative to act1+. his3+ mRNA was isolated from highly transcribed (strong nmt1 allele, no thiamine) and weakly transcribed conditions (weak nmt1 allele, thiamine) in swi6- ura4::L5-7kb::(Pnmt1-his3+)-ade6+ strains. For comparison the level of ade6+ and spcc330.03+ mRNA are shown for ura4::ade6+ strains. (B) Relative H3K9me2 enrichment for strongly and weakly transcribed conditions.
(2.72 MB TIF)
The ratio of Swi6p/H3K9me2 is reduced over spacer DNA. Scatter plot of the levels of Swi6p/H3K9me2 for heterochromatic regions genomewide [28], or within de novo heterochromatin domains.
(1.47 MB TIF)
Strains used in this study.
(0.04 MB PDF)
Primers used in this study.
(0.08 MB PDF)
Acknowledgments
We wish to thank Shiv Grewal for the swi6+333 strains and the anti-Swi6p antibody, Takeshi Urano for the anti-H3K9me2 antibody, Jian-Qiu Wu for the pFA6a series plasmids, Terilyn Gaither for technical assistance, and Laura Rusche for helpful discussion and critical reading of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported in part by an award from the Howard Hughes Medical Institute Professors Program (to HFW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 2.Schultz J. Variegation in Drosophila and the Inert Chromosome Regions. Proc Natl Acad Sci U S A. 1936;22:27–33. doi: 10.1073/pnas.22.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, et al. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 4.Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 5.Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedram M, Sprung CN, Gao Q, Lo AW, Reynolds GE, et al. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol Cell Biol. 2006;26:1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pravtcheva DD, Wise TL, Ensor NJ, Ruddle FH. Mosaic expression of an Hprt transgene integrated in a region of Y heterochromatin. J Exp Zool. 1994;268:452–468. doi: 10.1002/jez.1402680606. [DOI] [PubMed] [Google Scholar]
- 8.Muller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22:299–334. [Google Scholar]
- 9.Locke J, Kotarski MA, Tartof KD. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988;120:181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, et al. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 11.Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am J Hum Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, et al. Developmental Study of Fragile X Syndrome Using Human Embryonic Stem Cells Derived from Preimplantation Genetically Diagnosed Embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 14.Otten AD, Tapscott SJ. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc Natl Acad Sci U S A. 1995;92:5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 17.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. Embo J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 19.Sun FL, Elgin SC. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 20.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 21.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 22.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 23.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 25.Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- 26.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 27.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 28.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 29.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 30.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 31.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 32.Ayoub N, Goldshmidt I, Cohen A. Position effect variegation at the mating-type locus of fission yeast: a cis-acting element inhibits covariegated expression of genes in the silent and expressed domains. Genetics. 1999;152:495–508. doi: 10.1093/genetics/152.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayoub N, Goldshmidt I, Lyakhovetsky R, Cohen A. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics. 2000;156:983–994. doi: 10.1093/genetics/156.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken beta-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. Embo J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott KC, White CV, Willard HF. An RNA Polymerase III-Dependent Heterochromatin Barrier at Fission Yeast Centromere 1. PLoS ONE. 2007;2:e1099. doi: 10.1371/journal.pone.0001099. doi:10.1371/journal.pone.0001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, et al. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 40.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 41.Zurlinden A, Schweingruber ME. Identification of a DNA element in the fission yeast Schizosaccharomyces pombe nmt1 (thi3) promoter involved in thiamine-regulated gene expression. J Bacteriol. 1997;179:5956–5958. doi: 10.1128/jb.179.18.5956-5958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, et al. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Deng H, Bao X, Lerach S, Girton J, et al. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development. 2006;133:229–235. doi: 10.1242/dev.02199. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 47.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 48.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 49.Brink MC, van der Velden Y, de Leeuw W, Mateos-Langerak J, Belmont AS, et al. Truncated HP1 lacking a functional chromodomain induces heterochromatinization upon in vivo targeting. Histochem Cell Biol. 2006;125:53–61. doi: 10.1007/s00418-005-0088-7. [DOI] [PubMed] [Google Scholar]
- 50.Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, et al. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White WM, Willard HF, Van Dyke DL, Wolff DJ. The spreading of X inactivation into autosomal material of an X;autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donze D, Kamakaka RT. Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays. 2002;24:344–349. doi: 10.1002/bies.10072. [DOI] [PubMed] [Google Scholar]
- 54.Ferrari S, Simmen KC, Dusserre Y, Muller K, Fourel G, et al. Chromatin domain boundaries delimited by a histone-binding protein in yeast. J Biol Chem. 2004;279:55520–55530. doi: 10.1074/jbc.M410346200. [DOI] [PubMed] [Google Scholar]
- 55.Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, et al. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2:124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekinger EA, Gross DS. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. Embo J. 1999;18:7041–7055. doi: 10.1093/emboj/18.24.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dutrow N, Nix DA, Holt D, Milash B, Dalley B, et al. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 62.Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 63.Bi X, Yu Q, Sandmeier JJ, Zou Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol Cell Biol. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Festenstein R, Sharghi-Namini S, Fox M, Roderick K, Tolaini M, et al. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat Genet. 1999;23:457–461. doi: 10.1038/70579. [DOI] [PubMed] [Google Scholar]
- 65.Fourel G, Lebrun E, Gilson E. Protosilencers as building blocks for heterochromatin. Bioessays. 2002;24:828–835. doi: 10.1002/bies.10139. [DOI] [PubMed] [Google Scholar]
- 66.Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 67.Straub T, Becker PB. DNA sequence and the organization of chromosomal domains. Curr Opin Genet Dev. 2008;18:175–180. doi: 10.1016/j.gde.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 68.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 69.Miller AP, Willard HF. Chromosomal basis of X chromosome inactivation: identification of a multigene domain in Xp11.21–p11.22 that escapes X inactivation. Proc Natl Acad Sci U S A. 1998;95:8709–8714. doi: 10.1073/pnas.95.15.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci U S A. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valley CM, Pertz LM, Balakumaran BS, Willard HF. Chromosome-wide, allele-specific analysis of the histone code on the human X chromosome. Hum Mol Genet. 2006;15:2335–2347. doi: 10.1093/hmg/ddl159. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Willard HF, Mukherjee S, Furey TS. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol. 2006;2:e113. doi: 10.1371/journal.pcbi.0020113. doi:10.1371/journal.pcbi.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carrel L, Park C, Tyekucheva S, Dunn J, Chiaromonte F, et al. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet. 2006;2:e151. doi: 10.1371/journal.pgen.0020151. doi:10.1371/journal.pgen.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 75.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 76.Siam R, Dolan WP, Forsburg SL. Choosing and using Schizosaccharomyces pombe plasmids. Methods. 2004;33:189–198. doi: 10.1016/j.ymeth.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 77.Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 78.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 79.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pidoux A, Mellone B, Allshire R. Analysis of chromatin in fission yeast. Methods. 2004;33:252–259. doi: 10.1016/j.ymeth.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 81.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Umeda K, Yoko-o T, Nakayama K, Suzuki T, Jigami Y. Schizosaccharomyces pombe gmd3(+)/alg11(+) is a functional homologue of Saccharomyces cerevisiae ALG11 which is involved in N-linked oligosaccharide synthesis. Yeast. 2000;16:1261–1271. doi: 10.1002/1097-0061(200010)16:14<1261::AID-YEA620>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 83.Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. Embo J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pei Y, Schwer B, Saiz J, Fisher RP, Shuman S. RNA triphosphatase is essential in Schizosaccharomyces pombe and Candida albicans. BMC Microbiol. 2001;1:29. doi: 10.1186/1471-2180-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ura4 genomic region is not enriched in H3K9me2 in the absence of L5, and exhibits varible nucleosome occupancy. (A) The ura4 locus is depicted, and assayed for the presence of H3K9me2 (black) and Swi6p (grey), in the absence of the L5 element. (B) Nucleosome occupancy was characterized using an antibody to the c-terminus of histone H3. The data are expressed relative to the nucleosome occupancy at the act1+ locus.
(3.00 MB TIF)
S. pombe intergenic and lambda spacer fragments do not recruit H3K9me2 in the absence of L5. H3K9me2 enrichment over S. pombe intergenic (A) and lambda (B) sequences in the absence of L5. The intergenic spacer DNA is in duplicate copies in the genome (at the ura4 locus as well as its endogenous locus) thus no enrichment is represented by a value of 2.
(1.88 MB TIF)
Heterochromatin spreading in the ura4 locus is unaffected by the presence of lambda spacer DNA. Analysis of H3K9me2 in the ura4 locus in ura4::L5-7kb-ade6+ strains (black) and ura4::L5-ade6+ strains. To facilitate comparison between the two strains H3K9me2 enrichment for the ura4::L5-ade6+ is depicted with a gap over the lambda insert.
(1.93 MB TIF)
Increased swi6+ copy number results in decreased ade6+ expression but does not alter expression of other genes within the de novo heterochromatin domain. Steady state mRNA levels are depicted relative to ura4::ade6+ strains for wild type strains (grey) and strains bearing 3 copies of swi6+ (black).
(1.59 MB TIF)
Insertion of a gene within lambda attenuates heterochromatin independent of the level of transcription. (A) Levels of steady state mRNA relative to act1+. his3+ mRNA was isolated from highly transcribed (strong nmt1 allele, no thiamine) and weakly transcribed conditions (weak nmt1 allele, thiamine) in swi6- ura4::L5-7kb::(Pnmt1-his3+)-ade6+ strains. For comparison the level of ade6+ and spcc330.03+ mRNA are shown for ura4::ade6+ strains. (B) Relative H3K9me2 enrichment for strongly and weakly transcribed conditions.
(2.72 MB TIF)
The ratio of Swi6p/H3K9me2 is reduced over spacer DNA. Scatter plot of the levels of Swi6p/H3K9me2 for heterochromatic regions genomewide [28], or within de novo heterochromatin domains.
(1.47 MB TIF)
Strains used in this study.
(0.04 MB PDF)
Primers used in this study.
(0.08 MB PDF)