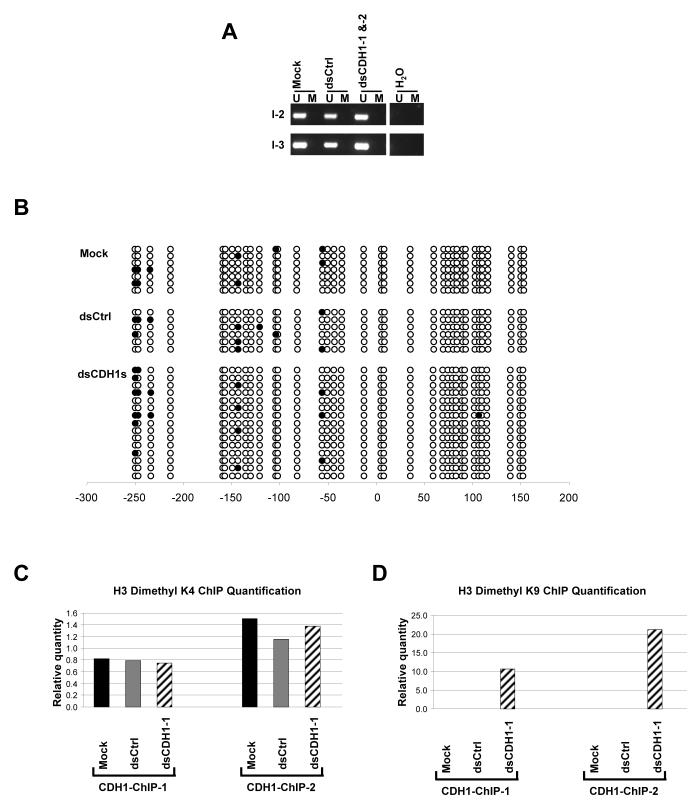
Figure 2.
DNA methylation and ChIP assay analyses on the CDH1 promoter after dsCDH1s treatment in HCT116 cells. (a) MSP analysis of the CpG island within the CDH1 promoter at 2 regions specified in Fig. 1a. The CDH1 promoter in HCT116 is normally unmethylated. DNA methylation patterns showing the presence of only the unmethylated (U) alleles remain unchanged in the mock, dsCtrl, and dsCDH1s treated cells. (b) Bisulfite sequencing analysis of the CpG island in the CDH1 promoter in the mock, dsCtrl, and dsCDH1s treated cells. The CpG sites are shown by their positions relative to the transcription start site (0), the open circles (○) represent unmethylated CpG sites, and closed circles (•) represent methylated CpG sites. The promoter remains unmethylated for each treatment group. (c) Real time PCR analysis of H3 dimethyl-K4 modification at the CDH1 promoter by ChIP assay. Two sets of primers were used in the PCR analysis to span the promoter as indicated in Fig. 1a. H3 dimethyl-K4 residues were observed, as expected, at both regions at the CDH1 promoter in the mock, dsCtrl, and dsCDH1-1 treated cells. (d) Real time PCR analysis of the H3 dimethyl-K9 modification at the CDH1 promoter by ChIP assay. H3 dimethyl-K9 was enriched at the CDH1 promoter in the dsCDH1-1 treated cells when compared to either the mock or the dsCtrl treated cells.