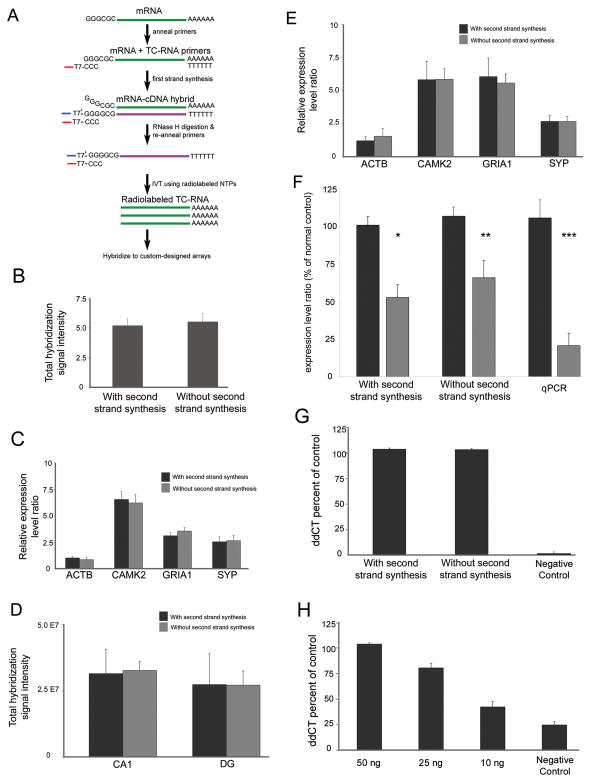
Figure 1. Comparison of TC RNA amplification methodologies.
A. Schematic representation of TC RNA amplification without second strand synthesis. The mRNA to be amplified (green line) and the TC primer serve as templates for the first strand synthesis with poly d(T) acting as a primer. First strand cDNA consists of three portions, the 5′ end comprised of the poly d(T), the mRNA complementary portion in the middle (purple line), and the 3′ end is comprised of the TC primer complementary to the cDNA (denoted as TC′). The TC′ portion hybridizes with the TC primer present in the reaction and forms a double stranded region without need for further second strand synthesis. Since the TC primer contains the T7 bacteriophage transcription promoter sequence, double stranded TC primer regions provide a functional RNA synthesis promoter for IVT and subsequent robust RNA amplification.
B. Histogram indicating no significant differences between the original TC RNA amplification method with second strand synthesis and TC RNA amplification without second strand synthesis in terms of total hybridization signal intensity ± standard deviation. Depicted assays were performed with 50 ng of mouse brain RNA using the original protocol (n = 6) and modified protocol (n = 8).
C. Comparison of amplification protocols using 50 ng of mouse brain RNA on specific gene expression levels. Histogram demonstrating no significant differences in expression levels in representative genes with varying levels of hybridization signal intensity. Key: ACTB, beta actin; CAMK2, calcium/calmodulin-dependent protein kinase II alpha; GRIA1, AMPA glutamate receptor 1 subunit; SYP, synaptophysin.
D. Similar to B, no differences in total hybridization signal intensity are observed between amplification protocols using RNA extracted from 50 cells per reaction obtained from normal control human postmortem CA1 neurons and DG granule cells.
E. Consistent with C, no differences are observed within representative individual genes following TC RNA amplification using the original or modified protocols in CA1 neurons acquired via LCM.
F. SYP expression levels are down regulated in CA1 neurons obtained from AD brains compared to age-matched normal controls using the original TC RNA amplification protocol (single asterisk denotes p < 0.01) and the TC RNA amplification without second strand synthesis methodology (double asterisk denotes p < 0.03). Consistent with the microarray analysis, significant down regulation of SYP expression was observed via qPCR in AD hippocampus as compared to age-matched controls (triple asterisk denotes p < 0.002).
G. Histograms depicting no significant differences in GRIA1 qPCR products across TC RNA amplification protocols in relation to a negative control using 50 ng of input mouse brain RNA.
H. GRIA1 qPCR products at three low concentrations (10 ng, 25 ng, and 50 ng) following TC RNA amplification without second strand synthesis. Note the linear nature of the amplification above the negative control.