Figure 2.
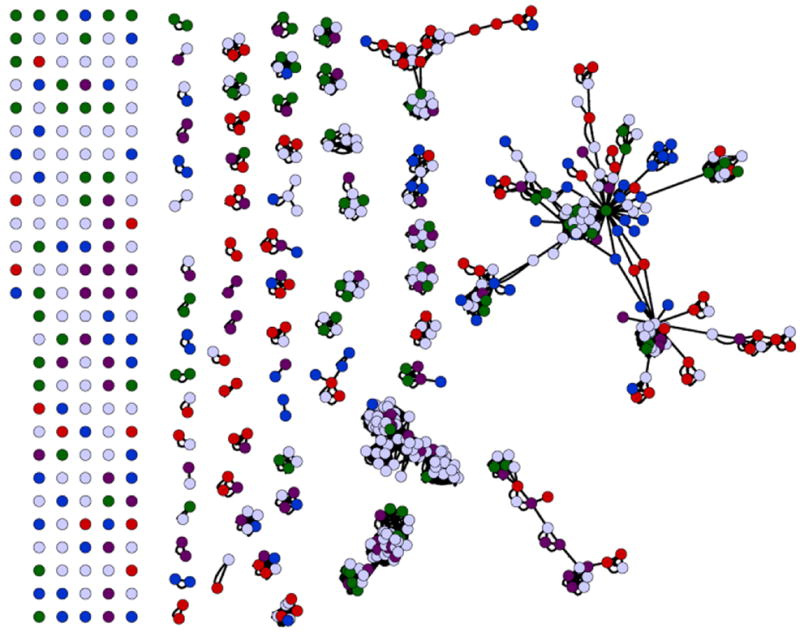
Genome-wide membrane protein target selection from S. cerevisiae for maximum Pfam coverage. The number of integral membrane proteins within yeast that have three or more TMH totals 621. Each protein is represented as a circle. These are clustered according to Pfam families. Colored circles represent the targets chosen for the expression of the first group of 384 membrane proteins, selected for maximal coverage of the Pfam families. As a result of clustering, there are 131 unannotated Pfam proteins, and 84 singletons that were all selected for expression (represented on the left side of the figure). 81 Pfam families have two or more members indicated by the clusters (center to right). In each case two members were chosen for remaining Pfam families. Seven additional proteins were selected from the larger clusters (right), to complete the target set of 384 membrane proteins. These proteins were divided into four sets of 96 tagets each (represented by red, blue, purple or green circles). Proximity within Pfams represents the sequence conservation between family members gleaned from use of multiple sequence alignment methods.
