Abstract
Perception and response to nutritional iron by bacteria is essential for viability, and for the ability to adapt to the environment. The iron response regulator (Irr) is part of a novel regulatory scheme employed by Rhizobium and other Alpha-Proteobacteria to control iron-dependent gene expression. Bradyrhizobium japonicum senses iron through the status of heme biosynthesis to regulate gene expression, thus it responds to an iron-dependent process rather than to iron directly. Irr mediates this response by interacting directly with ferrochelatase, the enzyme that catalyzes the final step in heme biosynthesis. Irr is expressed under iron limitation to both positively and negatively modulate gene expression, but degrades in response to direct binding to heme in iron-sufficient cells. Studies with Rhizobium reveal that the regulation of iron homeostasis in bacteria is more diverse than has been generally assumed.
INTRODUCTION
Iron bioavailability can be limiting because it is predominantly in the insoluble ferric form in aerobic environments. However, excessive intracellular iron can generate reactive oxygen species that damage cellular components (Braun and Killmann, 1999; Touati, 2000). Thus, iron homeostasis is strictly regulated so that iron acquisition, storage and consumption are geared to iron availability, and that intracellular levels of free iron do not reach toxic levels (reviewed in (Andrews et al., 2003)). In bacteria, the regulation of iron homeostasis has focused to a large extent on the Fur protein. The mechanism of Fur function has been particularly well studied in the Gamma-Proteobacteria E. coli (Crosa, 1997; Escolar et al., 1999; Hantke, 2001) and Pseudomonas aeruginosa (Ochsner and Vasil, 1996; Pohl et al., 2003), and in the gram-positive organism Bacillus subtilis (Baichoo and Helmann, 2002; Baichoo et al., 2002; Fuangthong and Helmann, 2003). Fur has been described primarily as a transcriptional repressor that binds DNA when bound to metal. However, direct activation of genes by Fur has been reported in Neisseria meningitidis (Delany et al., 2004) and a role for iron in attenuating DNA-binding activity was described for Helicobacter pylori Fur (Delany et al., 2001). Fur is the founding member of a family of proteins that are all involved in metalloregulation, but are specific for different regulatory metals (Bsat et al., 1998; Gaballa and Helmann, 1998; Hamza et al., 1998; Patzer and Hantke, 1998; Chao et al., 2004; Diaz-Mireles et al., 2004; Platero et al., 2004; Ahn et al., 2006)
Rhizobia are soil bacteria that can form a symbiotic relationship with leguminous plants. The bacteria infect roots of the plant host, leading to the development of root nodules. The intracellular bacteria fix atmospheric nitrogen to ammonia, which can be assimilated by the plant host to fulfill its nutritional nitrogen requirement. Rhizobia belong to the Alpha-Proteobacteria, an extremely diverse taxonomic group that includes pathogens, symbionts, photosynthetic organisms, bacteria that degrade environmental pollutants, and the abundant marine organism Pelagobacter ubique. The bacterial ancestor of mitochondria belongs to this group as well.
Recent studies show that iron metabolism is regulated very differently in the rhizobia compared to other well-studied model systems. Whereas Fur is the major global regulator of iron in E. coli, Pseudomonas aeruginosa and Bacillus subtilis, its role in Rhizobium is either diminished, has an alternative function or is absent altogether. In Sinorhizobium meliloti and Rhizobium leguminosarum, the Fur homolog is a manganese-responsive regulator and has been renamed Mur (Chao et al., 2004; Diaz-Mireles et al., 2004; Platero et al., 2004; Platero et al., 2007). The Fur protein from Bradyrhizobium japonicum is iron-responsive, but it has a diminished role in regulating iron transport genes and other genes controlled by Fur in E. coli, and it recognizes a novel DNA cis-acting element (Friedman and O’Brian, 2003, 2004; Yang et al., 2006c). Many of the regulatory functions ascribed to Fur in other bacteria are carried out by RirA or Irr in the rhizobia. RirA controls numerous iron-regulated genes, including those encoding proteins for iron transport, siderophore biosynthesis and iron-sulfur cluster assembly in S. meliloti and R. leguminosarum (Todd et al., 2002; Yeoman et al., 2004; Chao et al., 2005; Viguier et al., 2005).. RirA negatively controls gene expression in the presence of iron, and a RirA-responsive element is found in the promoters of genes under its control (Yeoman et al., 2004). rirA gene homologs are found in the Rhizobiaceae but not the Bradyrhizobiaceae
Irr protein belongs to the Fur family of metalloregulators that includes Fur, PerR, Zur, Nur and Mur (Bsat et al., 1998; Gaballa and Helmann, 1998; Hamza et al., 1998; Patzer and Hantke, 1998; Chao et al., 2004; Diaz-Mireles et al., 2004; Platero et al., 2004; Ahn et al., 2006). However, Irr behaves differently than these and other regulatory proteins in fundamentally different ways, and allows novel control of iron metabolism. Here, an overview of our current understanding of Irr function is reviewed.
Irr is a conditionally stable protein that degrades in response to iron in a heme-dependent manner
The irr gene was initially identified in Bradyrhizobium japonicum in a screen for loss of control of heme biosynthesis by iron (Hamza et al., 1998), and it has been most extensively characterized in that organism. Heme is the end product of a biosynthetic pathway, culminating with the insertion of iron into a protoporphyrin ring to produce protoheme. Irr coordinates the heme biosynthetic pathway with iron availability to prevent the accumulation of toxic porphyrin precursors under iron limitation (Hamza et al., 1998). Loss of function of the irr gene is sufficient to uncouple the pathway from iron-dependent control, as discerned by the accumulation of protoporphyrin. This accumulation is due to derepression of hemB and probably hemA (Hamza et al., 1998; Yang et al., 2006b). Similarly, an irr mutant of Rhizobium leguminosarum has a fluorescent colony phenotype and is deregulated for the hemA gene (Wexler et al., 2003; Todd et al., 2006), and a Brucella abortus irr mutant accumulates protoporphyrin (Martinez et al., 2005).
The Irr protein accumulates in cells under iron limitation, with very low levels in iron replete cells. Thus, Irr is distinct from other Fur family proteins because it functions in the absence of the regulatory metal, whereas the other members require direct metal-binding for activity. The control of iron on irr expression is primarily post-translational; Irr is a conditionally stable protein that degrades in cells exposed to iron (Qi et al., 1999). B. japonicum Irr contains a heme-regulatory motif (HRM) near the N-terminus that binds heme and is necessary for rapid degradation. Accordingly, Irr is stabilized in a heme-deficient background or by mutagenesis of cysteine-29 within the HRM.
Heme-mediated degradation is also found in other systems
Since the discovery of heme-dependent degradation of Irr, numerous other eukaryotic proteins have been identified that degrade in response to heme by binding to HRM motifs (Jeong et al., 2004; Ishikawa et al., 2005; Zenke-Kawasaki et al., 2007; Hu et al., 2008; Yang et al., 2008) (Table 1). The human iron regulatory protein 2 (IRP2) likely has a similar function as Irr in that the heme to which it responds reflects iron levels (Jeong et al., 2004; Ishikawa et al., 2005). Bach1 represses the transcription of genes encoding heme oxygenase, which uses heme as a substrate, and globins, which use heme as a prosthetic group. Thus, degradation in response to heme (Zenke-Kawasaki et al., 2007) coordinates heme availability with proteins that use it. The circadian clock in mammals is coordinated with heme biosynthesis such that the gene encoding the heme biosynthesis enzyme δ-aminolevulinic acid synthase is transcriptionally controlled (Zheng et al., 2001; Kaasik and Lee, 2004). Several regulators of the circadian clock have been identified and shown to bind heme (Dioum et al., 2002; Raghuram et al., 2007; Yin et al., 2007; Yang et al., 2008). One of them, Per2, contains an HRM that binds heme to affect its stability (Yang et al., 2008). Arginyl transferase catalyzes the addition of arginine to the N-terminal end of target proteins, which results in their degradation due to the N-end rule (Hu et al., 2008). Heme binds to arginyl transferase to inactivate it and to trigger its degradation by the proteasome. Interestingly, the ubiquitin ligase that tags proteins for degradation by covalent attachment of ubiquitin is also inhibited by heme (Hu et al., 2008), hence heme stabilizes proteins susceptible to arginylation by two separate mechanisms.
TABLE 1.
PROTEINS DEGRADED IN RESPONSE TO HRM BINDING BY HEME
| Protein | Cellular Process | Reference |
|---|---|---|
| Irr | Global iron homeostasis | Qi et al., 1999 |
| IRP2 | Global iron homeostasis | Ishikawi et al. 2005 |
| Bach1 | Globin and heme oxygenase regulation | Zenke-Kawasaki, 2007 |
| Arginyl transferase | N-end rule protein degradation | Hu et al., 2008 |
| Per2 | Circadian clock | Yang et al., 2008 |
Regulated degradation of B. japonicum Irr requires both redox states of heme
B. japonicum Irr fused to glutathione S transferase (GST) confers iron-dependent instability on GST, but a GST fusion containing only the N-terminal 36 amino acids of Irr, which includes the HRM, is stable (Yang et al., 2005). This means that the HRM is necessary but not sufficient for rapid degradation of Irr. In vitro and in vivo studies identified an instability domain that includes three consecutive histidines at positions 117–119, with His-117 and His-119 being invariant residues in Irr proteins. This domain is part of a heme-binding region distinct from the HRM. Whereas the HRM binds specifically to ferric (Fe3+) heme, the histidine-rich domain binds ferrous (Fe2+) heme (Yang et al., 2005). An Irr mutant in which the three histidines are replaced by alanines is stable in vivo under iron replete conditons (Yang et al., 2005). Irr decay follows first order kinetics (Qi et al., 1999), indicating a single mechanism for degradation. Hence the two hemes likely participate in a single degradation process rather than independent processes that occur at different rates. These findings implicate a role for the redox activity of heme in Irr degradation, and further evidence suggests that this activity leads to protein oxidation (Yang et al., 2006a). B. japonicum Irr degrades in response to cellular oxidative stress by a mechanism that involves heme and iron (Yang et al., 2006a). Furthermore, Irr degradation is strictly O2-dependent in vivo (Yang et al., 2006a). Irr oxidation was demonstrated in vitro, requiring heme, O2 and a reductant. An Irr truncation that does not bind ferrous heme in vitro does not degrade in vivo. Thus, it was proposed that reactive oxygen species participate in Irr degradation not only as part of an oxidative stress response (see below), but also in normal degradation in response to iron. Protein oxidation can result in hydrolysis of peptide bonds (Berlett and Stadtman, 1997) and thus, in principle, oxidation of Irr could be sufficient for degradation. However, in vivo degradation of Irr is rapid whereas carbonylation in vitro is slow. It is probable that oxidized Irr is recognized by cellular proteases as a damaged protein that is subsequently degraded. A candidate protease has not been described thus far. Degradation of both IRP2 and Bach1 require the ubiquitin ligase HOIL-1, which interacts with the heme-bound form of the respective protein (Ishikawa et al., 2005; Zenke-Kawasaki et al., 2007). The ubiquitin-tagged protein is then degraded. Arginyl transferase is tagged by N-end rule ubiquitin ligases in yeast and mouse for heme-dependent degradation (Hu et al., 2008).
Redox-dependent ligand switching, although not associated with protein degradation, occurs with the transcriptional regulator CooA from Rhodospirillum rubrum (Roberts et al., 2004) and the redox sensor EcDos from E. coli (Kurokawa et al., 2004). IRP2 binds ferric heme at the HRM and ferrous heme at a conserved histidine residue (Jeong et al., 2004; Ishikawa et al., 2005), and protein oxidation is heme-dependent (Yamanaka et al., 2003). Thus, there are similarities between IRP2 and Irr.
Examination of the Irr homologs reveals that only homologs within the Bradyrhizobiaceae have the Cys-Pro sequence and an HRM-like domain, whereas His-117 and His-119 of B. japonicum Irr are completely conserved in all of the homologs. This raises the question of whether Irr degradation as described for B. japonicum occurs in other related bacteria. A B. japonicum Irr derivative lacking an HRM degrades, but the rate is much slower than is found for the wild type protein (Yang et al., 2005). In addition, B. japonicum has a lower affinity ferric heme binding site (Qi and O’Brian, 2002) that could possibly serve a similar function as the HRM, albeit less efficiently. By analogy, Irr homologs lacking an HRM may have a compensatory mechanism that allows turnover.
Oxidative stress promotes degradation of the Irr protein
Bacteria have multiple defense strategies against oxidative stress, including the direct detoxification of ROS by catalase, peroxidases and superoxide dismutase. Oxidative stress responses require the activation of regulatory proteins and the induction of genes under their control. In many bacteria, the transcriptional regulator OxyR (Christman et al., 1989; Tao et al., 1989) senses hydrogen peroxide (Zheng et al., 1998) and induces numerous genes whose products are involved in peroxide defense (Tartaglia et al., 1989; Altuvia et al., 1994), redox balance (Prieto-Alamo et al., 2000; Ritz et al., 2000) and other factors (Altuvia et al., 1997; Zheng et al., 1999). In Bacillus subtilis, PerR is the major peroxide regulator and represses a large PerR regulon (Herbig and Helmann, 2001). The OhrR family of antioxidant regulators is responsible for organic hydroperoxide resistance (Mongkolsuk et al., 1998). B. japonicum contains an OxyR homolog, but it may function differently in that organism than in other systems (Panek and O’Brian, 2004). Evidence points to Irr as an oxidiative stress response regulator. Irr degrades in response to H2O2 produced endogenously in a catalase-deficient (katG) strain, or to H2O2 applied exogenously to culture media (Yang et al., 2006a). A Brucella abortus irr mutant displays elevated catalase activity, and resistance to killing by H2O2 (Martinez et al., 2006). The Irr deficiency causes derepression of hemB in B. japonicum and elevated heme in B. abortus. Catalases and peroxidases are heme proteins that detoxify H2O2 and peroxides, respectively, and elevated hemB may contribute to the synthesis of those enzymes. Other examples of elevated expression of heme biosynthesis genes are noted. The Bacillus subtilis PerR protein mediates the induction of the hemAXCDBL operon encoding enzymes for the early steps of heme synthesis (Chen et al., 1995; Mongkolsuk and Helmann, 2002). In E. coli and Salmonella, the hemH geneencoding the heme biosynthetic enzyme ferrochelatase is induced in response to H2O2 in an OxyR-dependent manner (Zheng et al., 2001; Elgrably-Weiss et al., 2002). However, it has not been established in B. japonicum or any other bacterium that synthesis of catalase or peroxidase substantially increases the overall heme demand in the cell, and thus the physiological relevance of elevated heme synthesis genes is uncertain.
Irr responds to heme at the site of heme synthesis, not to free heme
A fundamental problem with heme as a signaling molecule is that it is reactive and lipophilic. Heme can catalyze the formation of reactive oxygen species, and binds non-specifically to lipids, proteins and other macromolecules. Thus, a regulatory free heme pool is unlikely. The discovery of new and novel roles for heme as a regulatory molecule in eukaryotes and prokaryotes begs for reconciliation between these functions and the cytotoxicity of heme. This problem has been partially resolved for the Irr protein from B. japonicum. Ferrochelatase catalyzes the insertion of iron into protoporphyrin to form heme in the final step of the heme biosynthetic pathway. Irr interacts directly with ferrochelatase and responds to iron via the status of heme and protoporphyrin localized at the site of heme synthesis (Qi and O’Brian, 2002). Competition of the wild type ferrochelatase with a catalytically inactive one inhibits iron-dependent degradation of Irr even though the cell is not heme-defective. This means that Irr does not respond to a free heme pool, but rather to heme locally where it is synthesized. The dissociation binding constant (Kd) of heme for Irr is about 1 nM, which is less than one free heme molecule per cell. Irr may represent the simplest type of of heme signaling mechanism because there is no obvious need for a factor to chaperone heme from the site of synthesis to its target.
The interaction of Irr with ferrochelatase is affected by the immediate heme precursor protoporphyrin. The porphyrin-bound enzyme does not bind to Irr, which is the state of ferrochelatase when iron is limiting, and allows Irr to be active and affect the genes under its control. Thus, Irr is affected by heme and by its substrates so that heme synthesis does not exceed iron availability. In the presence of iron, ferrochelatase inactivates Irr, followed by Irr degradation to derepress the pathway. Irr is present but inactive in cells that express a catalytically inactive ferrochelatase, but active in a hemH deletion strain (Qi and O’Brian, 2002). It is possible that inactivation of Irr allows loss of function that is faster than its degradation. Indeed, the hemB mRNA is elevated by iron more rapidly than Irr degrades (Chauhan et al., 1997; Qi et al., 1999).
Irr is a global regulator of iron homeostasis
Although Irr was initially described in the context of heme biosynthesis, it is now clear that Irr is a global regulator of iron homeostasis and metabolism (Rudolph et al., 2006; Todd et al., 2006; Yang et al., 2006b). Irr was initially discovered as a positive effector of ferric iron transport as well as a negative regulator of heme biosynthesis (Hamza et al., 1998), and similar roles have been described for Brucella abortus Irr (Martinez et al., 2005, 2006). Microarray analysis of a B. japonicum irr mutant shows that Irr affects the expression of many iron-regulated genes in a positive and negative manner. In addition, several iron-regulated genes in R. leguminosarum are derepressed in an irr strain (Todd et al., 2006). However, iron transport is controlled by RirA in R. leguminosarum and S. meliloti, and thus the extent of the Irr regulon may be more limited in those species. A cis-acting DNA element called an iron control element (ICE) was found in the promoters of the divergently transcribed genes hmuR and hmuT from B. japonicum, and shown to be necessary for activation of those genes under iron limitation (Nienaber et al., 2001). This element was found to bind Irr in a yeast one hybrid screen (Rudolph et al., 2006), and hmuR and hmuT promoter activity is attenuated in an irr mutant (Nienaber et al., 2001). Furthermore, bioinformatic analyses identified ICE-like motifs upstream of many open reading frames in B. japonicum and other α-Proteobacteria (Rodionov et al., 2006; Rudolph et al., 2006). Evidence for control by Irr in the absence of an ICE motif was described in Brucella abortus (Martinez et al., 2006) and Bartonella quintana (Battisti et al., 2007).
Although Irr activity has been characterized extensively in the context of its negative control of hemB, that gene does not contain an ICE motif, and direct control by Irr remains unknown. However, in vitro Irr binds to ICE motifs in the promoters of blr7895 and the bacterioferritin gene bll6680, genes that are downregulated under iron limitation (Rudolph et al., 2006), consistent with a repressor role for Irr. Moreover, blr7895 and bll6680 are derepressed in an irr mutant, and Irr occupies the promoters of those genes in vivo (Sangwan et al., 2008). In addition, Irr represses transcription from the blr7895 promoter in vitro (Sangwan et al., 2008). Thus, Irr is a transcriptional repressor. Irr activity requires the presence of divalent metal in vitro for high affinity DNA binding, although the role of the metal is unknown (Sangwan et al., 2008).
Iron homeostasis is controlled by the status of heme via Irr
Irr interacts directly with the heme biosynthesis enzyme ferrochelatase, resulting in degradation under iron replete conditions, or accumulation of active protein under iron limitation (Qi and O’Brian, 2002). Thus, the discovery that Irr is a global regulator of iron-regulated genes indicates that iron homeostasis is controlled by the status of heme. Indeed, a heme-deficient strain of B. japonicum cannot maintain normal iron homeostasis. Control of Irr-regulated genes is aberrant in a heme-defective B. japonicum mutant, resulting in iron replete cells behaving as if they are iron-limited (Yang et al., 2006b). The heme mutant has an abnormally high cellular iron content, probably because iron transport genes are constitutively activated due to persistence of Irr in that strain. Accordingly, under iron limitation an irr mutant behaves as if it were iron replete even though cellular iron levels are lower than that found in the wild type (Yang et al., 2006b).
Most bacteria studied to date sense and respond to iron directly to regulate gene expression. That is, iron binds directly to a regulatory protein to modulate its activity. Iron binding to Fur confers DNA-binding activity on the protein, as also occurs for the DtxR regulator from Corynebacterium diphtheriae and the IdeR protein from Mycobacterium tuberculosis (Escolar et al., 1999; Pohl et al., 1999, 1999). However, B. japonicum, and perhaps other Alpha-proteobacteria, do not sense iron directly, but rather sense and respond to an iron-dependent process, namely the biosynthesis of heme. Is there an advantage to this type of control? Approximately one-half of the total iron in iron-limited B. japonicum cells is found in heme (unpublished observations). Since heme biosynthesis places such a high energy demand on the cell, this synthesis may serve as a sensitive indicator of the overall iron status. Also, many iron-dependent processes such as electron transport, tricarboxylic acid cycle and detoxification are associated with aerobic metabolism, which also requires heme. Therefore, it may allow a better coordination of cellular events.
Taxonomic distribution of Irr
Irr is prevalent in the Alpha subdivision of the proteobacterial phylum. Amongst the sequenced genomes, it is ubiquitious in the order Rhizobiales and Rhodobacteriales, and found in some Rhodospirilles as well (reviewed in (Rodionov and Gelfand, 2006). It is also present in the marine bacterium Pelagibacter ubique, which is in the order Rickettsiales, but is not present in its obligate intracellular relatives Rickettsia, Wolbachia or Ehrlichia. Interestingly, an Irr homolog is also found in Acidothiobacillus ferrooxidans, a Gamma-Proteobacterium that lives in acidic environments and is exposed to iron predominantly in the ferrous form. Microarray analysis shows that the vast majority of B. japonicum genes that are strongly regulated by iron are under the control of Irr. Thus, Irr is the major iron regulator in that bacterium and probably in other Bradyrhizobiaceae, with Fur having a much less prominent role. However, many rhizobia contain RirA in addition to Irr (Todd et al., 2002; Yeoman et al., 2004; Chao et al., 2005; Todd et al., 2005; Viguier et al., 2005). Numerous cellular systems controlled by Irr in B. japonicum are regulated by RirA in S. meliloti and R. leguminosarum (Todd et al., 2002; Chao et al., 2005; Viguier et al., 2005). Notably, heme and ferric iron transport genes are controlled by RirA in those two species, but by Irr in B. japonicum. These systems are negatively controlled by RirA in the presence of iron in species, whereas Irr appears to positively control them under low iron conditions. Brucella abortus contains both Irr and RirA, but siderophore genes in that organism are controlled by Irr (Martinez et al., 2006), therefore it may not be possible to assume a priori the role of RirA in organisms that contain it. Nevertheless, Irr and RirA appear to usurp the role of Fur to varying degrees in organisms that contain these novel regulators.
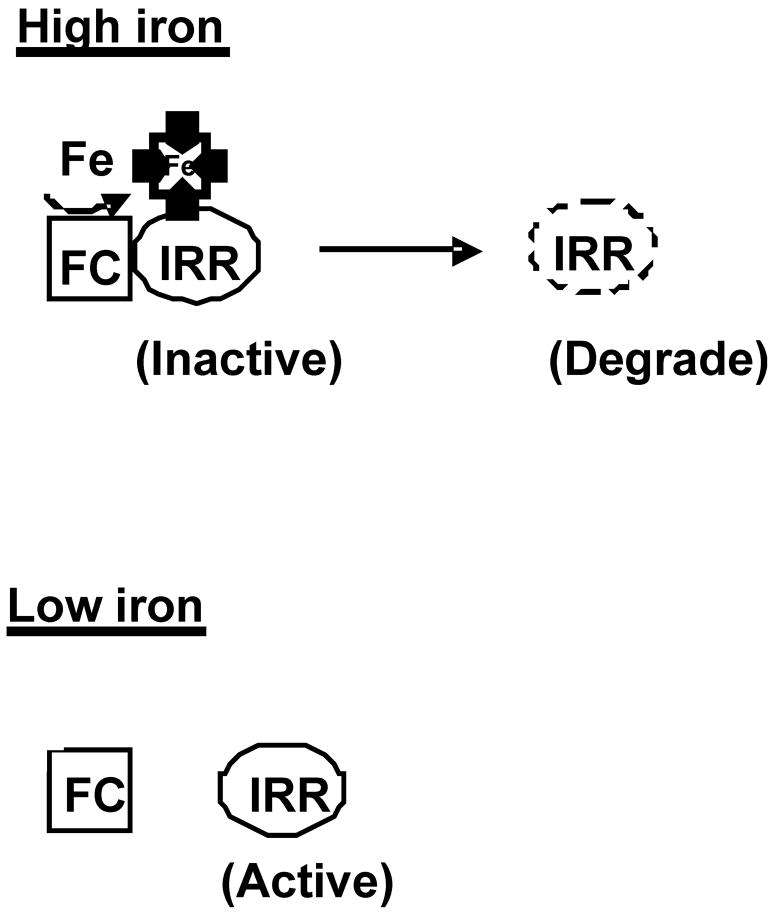
FIGURE 1. Regulation of Irr activity and degradation at the site of heme synthesis.
FC denotes ferrochelatase. The light porphyrin represents protoporphyrin and the dark porphyrin with the inserted iron molecule is heme (protoheme). Irr responds to heme at the site of heme synthesis rather than responding to a free heme pool. Irr inactivation precedes its degradation.
Acknowledgments
We thank former and current members of the O’Brian lab. This work is supported by National Institutes of Health grant GM-067966 to M.R.O’B
References
- Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184:5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- Battisti JM, Smitherman LS, Sappington KN, Parrow NL, Raghavan R, Minnick MF. Transcriptional regulation of the heme binding protein gene family of Bartonella quintana is accomplished by a novel promoter element and iron response regulator. Infect Immun. 2007;75:4373–4385. doi: 10.1128/IAI.00497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologs: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Chao TC, Becker A, Buhrmester J, Puhler A, Weidner S. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol. 2004;186:3609–3620. doi: 10.1128/JB.186.11.3609-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao TC, Buhrmester J, Hansmeier N, Puhler A, Weidner S. Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl Environ Microbiol. 2005;71:5969–5982. doi: 10.1128/AEM.71.10.5969-5982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Titus DE, O’Brian MR. Metals control activity and expression of the heme biosynthesis enzyme δ-aminolevulinic acid dehydratase in Bradyrhizobium japonicum. J Bacteriol. 1997;179:5516–5520. doi: 10.1128/jb.179.17.5516-5520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Keramati L, Helmann JD. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman MF, Storz G, Ames BN. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol. 2004;52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- Delany I, Spohn G, Rappuoli R, Scarlato V. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol Microbiol. 2001;42:1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AW. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology. 2004;150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Elgrably-Weiss M, Park S, Schlosser-Silverman E, Rosenshine I, Imlay J, Altuvia S. A Salmonella enterica serovar typhimurium hemA mutant is highly susceptible to oxidative DNA damage. J Bacteriol. 2002;184:3774–3784. doi: 10.1128/JB.184.14.3774-3784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman YE, O’Brian MR. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J Biol Chem. 2003;278:38395–38401. doi: 10.1074/jbc.M306710200. [DOI] [PubMed] [Google Scholar]
- Friedman YE, O’Brian MR. The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J Biol Chem. 2004;279:32100–32105. doi: 10.1074/jbc.M404924200. [DOI] [PubMed] [Google Scholar]
- Fuangthong M, Helmann JD. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J Bacteriol. 2003;185:6348–6357. doi: 10.1128/JB.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I, Chauhan S, Hassett R, O’Brian MR. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J Biol Chem. 1998;273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Herbig AF, Helmann JD. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol. 2001;41:849–859. doi: 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- Hu RG, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kato M, Hori H, Ishimori K, Kirisako T, Tokunaga F, Iwai K. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol Cell. 2005;19:171–181. doi: 10.1016/j.molcel.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Jeong J, Rouault TA, Levine RL. Identification of a heme-sensing domain in iron regulatory protein 2. J Biol Chem. 2004;279:45450–45454. doi: 10.1074/jbc.M407562200. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Lee DS, Watanabe M, Sagami I, Mikami B, Raman CS, Shimizu T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–20193. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
- Martinez M, Ugalde RA, Almiron M. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology. 2005;151:3427–3433. doi: 10.1099/mic.0.28213-0. [DOI] [PubMed] [Google Scholar]
- Martinez M, Ugalde RA, Almiron M. Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology. 2006;152:2591–2598. doi: 10.1099/mic.0.28782-0. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienaber A, Hennecke H, Fischer HM. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol Microbiol. 2001;41:787–800. doi: 10.1046/j.1365-2958.2001.02555.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Vasil ML. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek HR, O’Brian MR. KatG is the primary detoxifier of hydrogen peroxide produced by aerobic metabolism in Bradyrhizobium japonicum. J Bacteriol. 2004;186:7874–7880. doi: 10.1128/JB.186.23.7874-7880.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Platero R, de Lorenzo V, Garat B, Fabiano E. Sinorhizobium meliloti fur-like (Mur) protein binds a fur box-like sequence present in the mntA promoter in a manganese-responsive manner. Appl Environ Microbiol. 2007;73:4832–4838. doi: 10.1128/AEM.00686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero R, Peixoto L, O’Brian MR, Fabiano E. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl Environ Microbiol. 2004;70:4349–4355. doi: 10.1128/AEM.70.7.4349-4355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- Pohl E, Holmes RK, Hol WG. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J Mol Biol. 1999;292:653–667. doi: 10.1006/jmbi.1999.3073. [DOI] [PubMed] [Google Scholar]
- Pohl E, Holmes RK, Hol WG. Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied. J Mol Biol. 1999;285:1145–1156. doi: 10.1006/jmbi.1998.2339. [DOI] [PubMed] [Google Scholar]
- Prieto-Alamo MJ, Jurado J, Gallardo-Madueno R, Monje-Casas F, Holmgren A, Pueyo C. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J Biol Chem. 2000;275:13398–13405. doi: 10.1074/jbc.275.18.13398. [DOI] [PubMed] [Google Scholar]
- Qi Z, Hamza I, O’Brian MR. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc Natl Acad Sci USA. 1999;96:13056–13061. doi: 10.1073/pnas.96.23.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, O’Brian MR. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol Cell. 2002;9:155–162. doi: 10.1016/s1097-2765(01)00431-2. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D, Patel H, Doan B, Zheng M, Aslund F, Storz G, Beckwith J. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J Biol Chem. 2000;275:2505–2512. doi: 10.1074/jbc.275.4.2505. [DOI] [PubMed] [Google Scholar]
- Roberts GP, Youn H, Kerby RL. CO-sensing mechanisms. Microbiol Mol Biol Rev. 2004;68:453–473. doi: 10.1128/MMBR.68.3.453-473.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS. Computational identification of BioR, a transcriptional regulator of biotin metabolism in Alphaproteobacteria, and of its binding signal. FEMS Microbiol Lett. 2006;255:102–107. doi: 10.1111/j.1574-6968.2005.00070.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational Reconstruction of Iron- and Manganese-Responsive Transcriptional Networks in alpha-Proteobacteria. PLoS Comput Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, Hennecke H, Fischer HM. The Iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J Bacteriol. 2006;188:733–744. doi: 10.1128/JB.188.2.733-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan I, Small SK, O’Brian MR. The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high affinity DNA-binding activity. J Bacteriol. 2008 doi: 10.1128/JB.00495-00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K, Makino K, Yonei S, Nakata A, Shinagawa H. Molecular cloning and nucleotide sequencing of oxyR, the positive regulatory gene of a regulon for an adaptive response to oxidative stress in Escherichia coli: homologies between OxyR protein and a family of bacterial activator proteins. Mol Gen Genet. 1989;218:371–376. doi: 10.1007/BF00332397. [DOI] [PubMed] [Google Scholar]
- Tartaglia LA, Storz G, Ames BN. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- Todd JD, Sawers G, Johnston AW. Proteomic analysis reveals the wide-ranging effects of the novel, iron-responsive regulator RirA in Rhizobium leguminosarum bv. viciae. Mol Genet Genomics. 2005;273:197–206. doi: 10.1007/s00438-005-1127-8. [DOI] [PubMed] [Google Scholar]
- Todd JD, Sawers G, Rodionov DA, Johnston AW. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol Genet Genomics. 2006;275:564–577. doi: 10.1007/s00438-006-0115-y. [DOI] [PubMed] [Google Scholar]
- Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AW. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology. 2002;148:4059–4071. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- Viguier C, Cuiv PO, Clarke P, O’Connell M. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol Lett. 2005;246:235–242. doi: 10.1016/j.femsle.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Wexler M, Todd JD, Kolade O, Bellini D, Hemmings AM, Sawers G, Johnston AW. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology. 2003;149:1357–1365. doi: 10.1099/mic.0.26130-0. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Ishikawa H, Megumi Y, Tokunaga F, Kanie M, Rouault TA, Morishima I, Minato N, Ishimori K, Iwai K. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat Cell Biol. 2003;5:336–340. doi: 10.1038/ncb952. [DOI] [PubMed] [Google Scholar]
- Yang J, Ishimori K, O’Brian MR. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J Biol Chem. 2005;280:7671–7676. doi: 10.1074/jbc.M411664200. [DOI] [PubMed] [Google Scholar]
- Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, Mury SP, Capelluto DG, Finkielstein CV. A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol Cell Biol. 2008;28:4697–4711. doi: 10.1128/MCB.00236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Panek HR, O’Brian MR. Oxidative stress promotes degradation of the Irr protein to regulate haem biosynthesis in Bradyrhizobium japonicum. Mol Microbiol. 2006a;60:209–218. doi: 10.1111/j.1365-2958.2006.05087.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer HM, O’Brian MR. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol. 2006b;60:427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Sangwan I, O’Brian MR. The Bradyrhizobium japonicum Fur protein is an iron-responsive regulator in vivo. Mol Genet Genomics. 2006c;276:555–564. doi: 10.1007/s00438-006-0162-4. [DOI] [PubMed] [Google Scholar]
- Yeoman KH, Curson AR, Todd JD, Sawers G, Johnston AW. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology. 2004;150:4065–4074. doi: 10.1099/mic.0.27419-0. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, Asahara T, Tokunaga F, Iwai K, Igarashi K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27:6962–6971. doi: 10.1128/MCB.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]