Fig. 3. Population genomics: variation and selection.
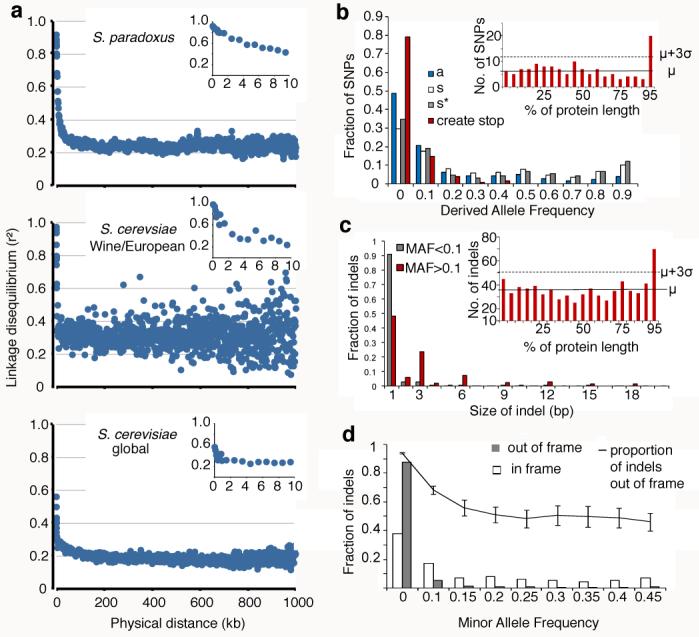
a, Linkage disequilibrium as a function of distance averaged over 1kb. Insets show the decline in linkage disequilibrium over the first 10kb. Details shown in Table S7.
b, Derived allele frequencies of SNPs in coding regions. Amino acid changing SNPs (‘a’) show an excess of low frequencies compared to synonymous SNPs (‘s’). Synonymous SNPs in genes with strong codon bias (‘s*’) are in excess at low and high frequencies. SNPs that create stop codons (‘create stop’) show skew to low frequencies. Inset is the number of mutations occurring over the length of the protein, exceeding three standard deviations from the mean in the C-terminus.
c, Distribution of sizes of indel polymorphisms in coding regions. High frequency indels (>10%, red) more often occur in multiples of 3 than low frequency indels (grey). Inset is as for b.
d, Frequency distribution of indels in coding regions. Out of frame indels (grey) show excess at low frequencies relative to in frame indels (unfilled). The proportion of out of frame indels decreases as frequency increases. Error bars represent the standard error of the proportion.
