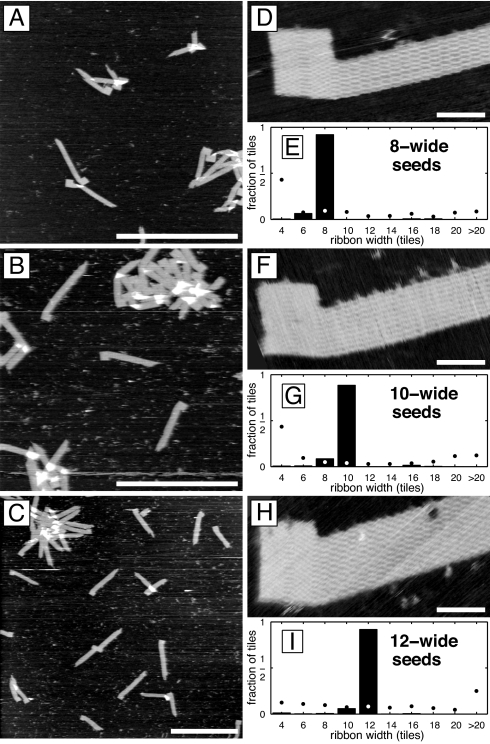
Fig. 3.
Programming ribbon width. (A–C) AFM images of ligated ribbons grown from seeds specifying 8-, 10-, and 12-tile-wide ribbons, respectively. (Scale bars: 1 μm.) (D, F, and H) Respective high-resolution AFM images of individual ribbons. (Scale bars: 50 nm.) (E, G, and I) Distribution of ribbon widths in corresponding samples of unligated ribbons (SI Text), given as the fraction of tiles found in ribbons of a given width. Solid bars indicate samples annealed with seeds. n = 41,718; 69,200; and 125,876 tiles, respectively. Dots indicate samples annealed without seeds. n = 23,524; 26,404; and 145,376 tiles, respectively. In each experiment, boundary and nucleation barrier tiles were at 100 nM each, and the repeatable block tile concentrations were proportional to their use in ribbons of the target width, i.e., 200, 300, and 400 nM, respectively. For samples with seeds, each staple strand was at 50 nM, each adapter strand was at 100 nM, and the origami scaffold strand was at 10 nM.