Fig. 2.
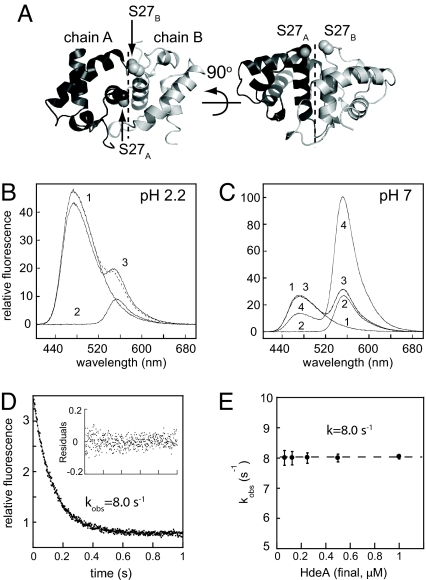
The oligomeric state of HdeA can be monitored by FRET. (A) Ribbon representation of the HdeA dimer [Protein Data Bank (PDB) ID code 1DJ8; chains A and B from reference 12] showing the position of S27, which was mutated to cysteine to provide a fluorescence-labeling site. (B) Fluorescence emission spectra of 0.5 μM bimane-labeled HdeA(C27) (trace 1), AF532-labeled HdeA(C27) (trace 2), or a 1:1 mixture of both (trace 3) recorded at pH 2.2 (λex = 390 nm). The sum of spectra 1 and 2 are shown as a dashed line. (C) Fluorescence emission spectra of 0.5 μM bimane-labeled HdeA(C27) (trace 1), AF532-labeled HdeA(C27) (trace 2), or a 1:1 mixture of both recorded at pH 7 (trace 3), or a 1:1 mixture of both mixed at pH 2.2 and then neutralized (trace 4). The sum of spectra 1 and 2 are shown as a dashed line, which fits nearly perfectly to trace 3 and is therefore not clearly visible. (D) Kinetics of pH-induced dimer dissociation as measured by a decrease in intermolecular FRET. Bimane- and AF532-labeled HdeA(C27) dimers were prepared by incubating 1:1 mixtures at low pH and subsequently diluting to a final concentration of 0.5 μM in pH 7 buffer. Dissociation of the dimers was then induced by rapidly shifting back to low pH by using a stopped-flow device. A representative trace is shown. (Inset) Residuals for a single exponential fit. (E) Plot of kobs over a range of HdeA concentrations. The dashed line denotes the average value of 8.0 s−1.