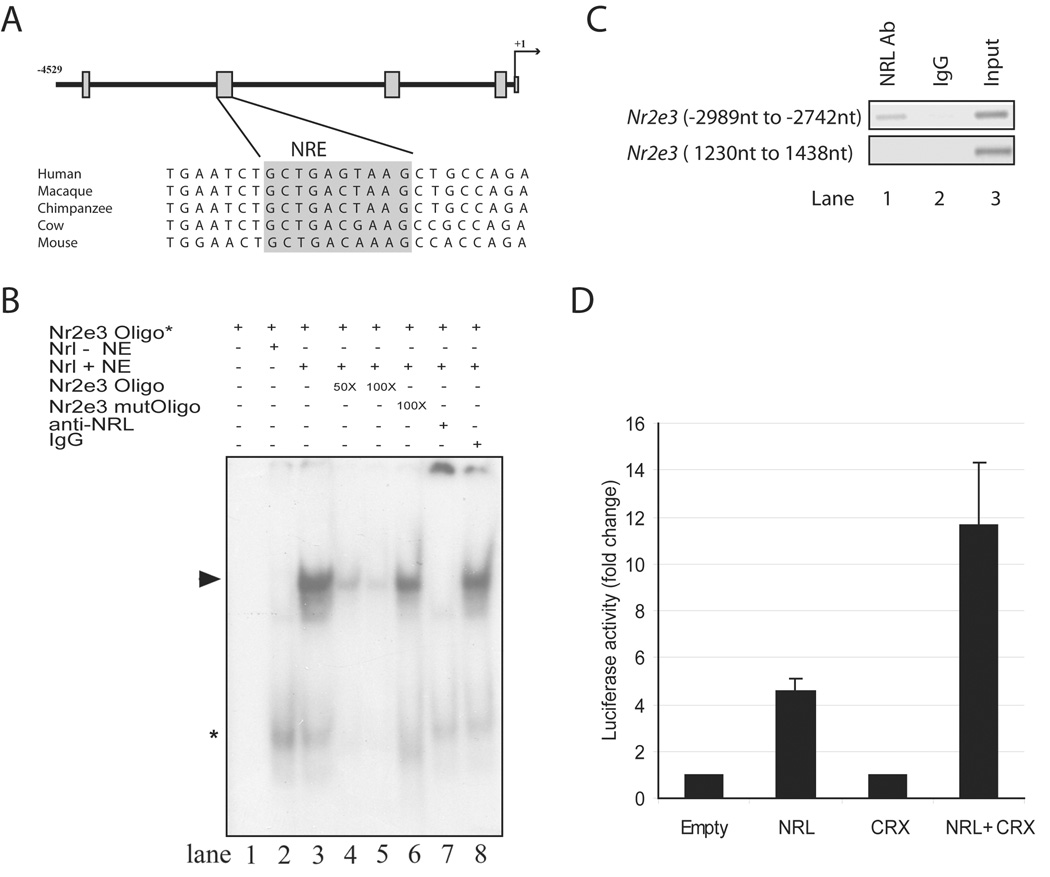
Figure 1. Binding to and activation of the Nr2e3 promoter by NRL.
(A) Schematic of approximately 4.5 kb genomic DNA upstream of the Nr2e3 transcription start site (denoted as +1). The four boxes indicate sequence regions conserved in mammals. A comparison of sequences in the second conserved region including a putative NRE (highlighted in grey) is shown below the schema. (B) EMSA showing the binding of NRL to NRE site in the Nr2e3 promoter. Lanes are as indicated above the autoradiograph. Nr2e3 oligo* indicates 32P-labeled NRE oligonucleotide (−2820 nt to −2786 nt) (in all lanes). Nrl – NE shows 10 µg nuclear extract from untransfected COS-1 cells (lane 2), whereas Nrl + NE means 10 µg nuclear extract from COS-1 cells transfected with Nrl cDNA expression plasmid (lanes 3–8). Lane 4 and 5 included 50- or 100 fold molar excess of unlabeled NRE oligonucleotide. Lane 6 included 100-fold molar excess of unlabeled mutant NRE oligonucleotide. Lane 7 contains 2 µg anti-NRL antibody, whereas lane 8 has 2 µg normal rabbit IgG. Arrowhead represents the specific shifted band, which is undetectable when anti-NRL antibody is included. Asterisk indicates a shifted band (of low molecular mass) that does not seem to be altered by the addition of anti-NRL antibody. These experiments were performed three times, and similar results were obtained. (C) PCR assays using immunoprecipitated chromatin from adult C57BL/6J retinas. Lane1, NRL antibody used for IP; lane 2, normal rabbit IgG used for IP (negative control); lane 3, input DNA used as template. Top panel: primers amplifying the NRE containing region (−2989 nt to −2742 nt) in the Nr2e3 promoter region were used for PCR. Bottom panel: primers amplifying an irrelevant region (1230 nt to 1438 nt) in the Nr2e3 gene were used for PCR. (D) Luciferase reporter assays showing the activation of Nr2e3 promoter by NRL and CRX.