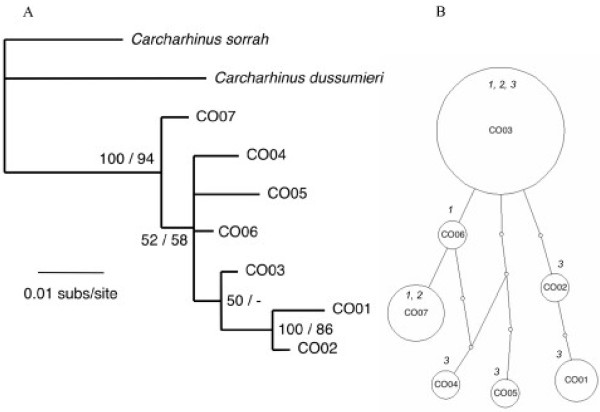
Figure 2.
Inferred phylogeny (A) and statistical parsimony network (B) among haplotypes of C. obscurus. The phylogeny was rooted with C. sorrah and C. dussumieri and nodal support is given as Bayesian posterior probabilities/ML boostrap support. Dash (-) indicates support of less than 50%. In the network, each indicated step (circle) represents a single nucleotide difference in the mtDNA control region sequence. The area of circles is scaled to represent the relative frequency of that haplotype and the smallest circle represent inferred haplotypes that were not sampled. The collection location of sampled haplotypes is numbered (in italics) according to Fig. 1.