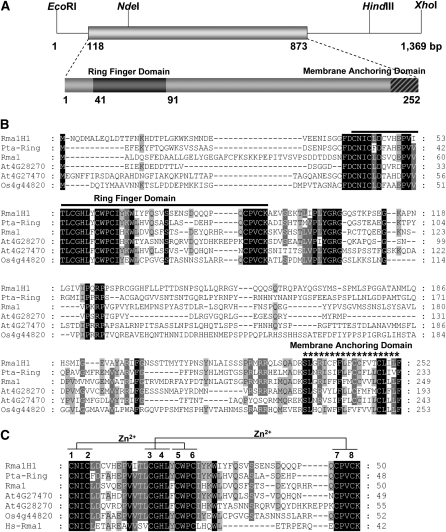
Figure 1.
Sequence Analysis of Hot Pepper Rma1H1.
(A) Restriction enzyme map analysis and schematic structure of the hot pepper Rma1H1 cDNA clone and predicted Rma1H1 protein. Solid bar represents the coding region. Solid lines depict 5′- and 3′-untranslated regions. Dark bar indicates N-terminal RING motif, while hatched bar shows C-terminal membrane anchoring domain.
(B) Comparison of the derived amino acid sequence of hot pepper Rma1H1 with those of the poplar Pta-Ring protein, Arabidopsis RING membrane anchor 1 (Rma1), Rma2 (At4g28270), and Rma3 (At4g27470) proteins, and rice RING (Os4g44820) protein. Amino acid residues that are conserved in at least four of the six sequences are shaded, while amino acids that are identical in all six proteins are shown in black. The solid line denotes the N-terminal RING motif, which is essential for E3 Ub ligase activity. The C-terminal putative membrane anchoring sequence is indicated by an asterisk. Dashes show gaps in the amino acid sequences that were introduced to optimize alignment.
(C) Sequence alignment of the RING domain of Rma1H1 and other RING proteins. The sequences of RING motifs in hot pepper Rma1H1, Arabidopsis Rma1, Rma2, and Rma3, poplar Pta-Ring protein, rice RING protein, and human Hs-Rma1 are shown. Amino acid residues that are conserved in at least four of the seven sequences are shaded. Amino acids that are identical in all seven proteins are shown in black. Putative Zn2+-interacting amino acid residues are indicated. The numbers on the right indicate the amino acid residues. Dashes show gaps in the amino acid sequences that were introduced to optimize alignment.