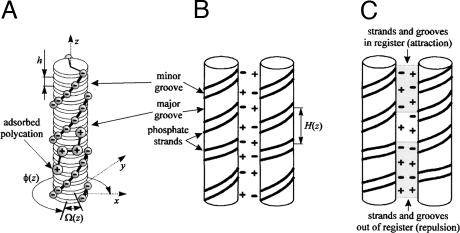
Fig. 2.
Accumulating mismatch (taken from ref. 16). (A) B-DNA sketched as a stack of base pairs (disks). Each base pair has two negatively charged phosphate groups. The base pair orientation at the altitude z is described by the azimuthal phase angle Φ(z) of the middle of the minor groove. Each combination of adjacent base pairs has a preferred twist angle Ω(z) = 〈Ω〉 = + ΔΩ(z), where 〈Ω〉 = 34–35° and = 4–6° (22). For rigid molecules the phase angle accumulates according to the preferred twist angles between adjacent base pairs, i.e., Φ(z) = ∫0z Ω(z′)dz′. The deviations of the phase angle from ideal helicity accumulate along the z axis as a “random walk” over a characteristic length, called the helical coherence length λc = h/(ΔΩ)2 [ΔΩ given in radians (16)], beyond which nonhomologous molecules cannot maintain favorable juxtaposition (B) The sequence-dependent twist modulation, Ω(z), leads to axial variation of the local helical pitch. As a result, only DNA with homologous sequences can have negatively charged strands facing positively charged grooves over a large juxtaposition length. (For visualization, the variation of H(z) is strongly exaggerated). (C) Molecules with unrelated sequences have uncorrelated twist modulations; this results in the loss of register between opposing strands and grooves, and a larger interaction energy. The loss of register takes place over the length λc, which determines the length of a sequence above which the double helices can sense the difference between the juxtaposition of homologous and nonhomologous tracks.