Abstract
IIAGlc, the glucose-specific phosphocarrier protein of the phosphoenolpyruvate:glycose phosphotransferase system, is an allosteric inhibitor of Escherichia coli glycerol kinase. A linked-functions initial-velocity enzyme kinetics approach is used to define the MgATP-IIAGlc heterotropic allosteric interaction. The interaction is measured by the allosteric coupling constants Q and W, which describe the mutual effect of the ligands on binding affinity and the effect of the allosteric ligand on Vmax, respectively. Allosteric interactions between these ligands display K-type activation and V-type inhibition. The allosteric coupling constant Q is about 3, showing cooperative coupling such that each ligand increases the affinity for binding of the other. The allosteric coupling constant W is about 0.1, showing that the allosteric inhibition is partial such that binding of IIAGlc at saturation does not reduce Vmax to zero. E. coli glycerol kinase is a member of the sugar kinase/heat shock protein 70/actin superfamily, and an element of the superfamily conserved ATPase catalytic core was identified as part of the IIAGlc inhibition network because it is required to transplant IIAGlc allosteric control into a non-allosteric glycerol kinase (Pawlyk, A.C. and Pettigrew, D.W. (2002) Proc. Natl. Acad. Sci. USA 99:11115-20). Two of the amino acids at this locus of E. coli glycerol kinase are replaced with those from the non-allosteric enzyme to enable determination of its contributions to MgATP-IIAGlc allosteric coupling. The substitutions reduce the affinity for IIAGlc by about 5-fold without changing significantly the allosteric coupling constants Q and W. The insensitivity of the allosteric coupling constants to the substitutions may indicate that the allosteric network is robust or the locus is not an element of that network. These possibilities may arise from differences of E. coli glycerol kinase relative to other superfamily members with respect to oligomeric structure and location of the allosteric site in a single domain far from the catalytic site.
Allosteric control of enzyme catalysis is a central aspect of regulation in biological systems. In central metabolic pathways, modulation of catalytic activity coordinates responses to changing carbon source availability, energy requirements, or needs for specific molecules for anabolism. Classical descriptions of allostery are based on global conformational change in response to binding of a regulatory ligand to its allosteric site that results in change in the conformation and function of the catalytic site [1]. Developing views of allostery differ in several respects from the classical description. Among the most important of these differences is the view that communication between the allosteric and catalytic sites is mediated by interactions in a sparse network of amino acid residues [2-7]. In this context, identification of the interacting residues and the roles of each residue in the allosteric communication become important objectives for experiments [8]. Approaches for identification of these networks include bioinformatics methods [7, 9, 10], computational methods [2, 4, 11-13], and structure-based experimental methods [5, 14-18]. We used an approach that is based on transplanting allosteric control into a naïve enzyme to identify a locus that is part of an allosteric network [19] and showed that the locus modulates local motions and binding affinity at the allosteric site [20]. Here, we evaluate the role of this locus in the coupling between the allosteric and catalytic sites.
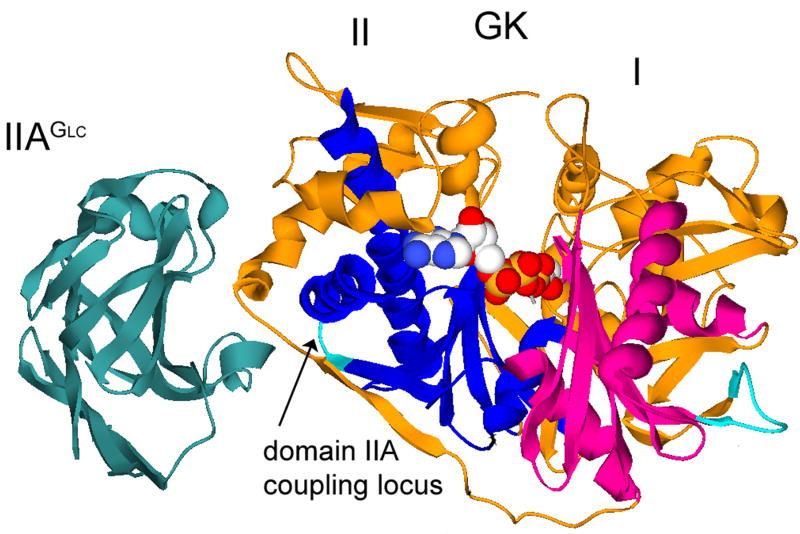
Catalytic activity of Escherichia coli glycerol kinase (EGK)1 is inhibited by the unphosphorylated form of IIAGlc, the glucose-specific phosphocarrier protein of the E. coli phosphoenolpyruvate:glycose phosphotransferase system. During steady-state glucose uptake, IIAGlc is about 95% unphosphorylated in vivo [21] and its inhibition of EGK and membrane systems for lactose, melibiose, and maltose uptake is believed to be a central mechanism by which glucose prevents metabolism of other carbon sources [22, 23]. The co-crystal structure of the EGK-IIAGlc complex [24] that is presented in figure 1 shows that EGK is a member of the sugar kinase/hsp70/actin superfamily [25]. ATPase catalytic activity is a shared function of superfamily members and the activities of many of the superfamily members are controlled allosterically. Functional properties of superfamily members can be considered in the context of a shared conserved structure. The shared structure of superfamily members has two domains, termed I and II. These domains are divided into subdomains and a conserved ATPase catalytic core is located in subdomains IA and IIA. The ATPase catalytic site is at the interface between domains I and II, and the EGK substrates interact with both domains (figure 1). Substrate binding to superfamily members is associated with closure of the catalytic site cleft [18, 24, 26-32] and allosteric control of the ATPase enzyme activity is generally believed to be mediated by modulation of this conformational change [25, 33]. For the superfamily members glucokinase, actin, and hsp70, crystal structures show that allosteric effectors interact with both domains and appear to exert direct steric effects on the conformational change [34-41]. The binding site for IIAGlc is on only domain II of EGK and is 30 Å from the catalytic site [24]. Thus, direct steric effects on the catalytic site closure do not appear to account for IIAGlc inhibition of EGK.
Figure 1.
Co-crystal structure of EGK-IIAGlc. Ribbon structures of EGK and IIAGlc are shown with labels for each protein and the domains of EGK. The conserved ATPase cores of subdomains IA and IIA are colored in magenta and blue, respectively. The coupling locus in subdomain IIA is colored in cyan and is located at the junction between two layers of the conserved ATPase core, as indicated by the labeled arrow. The substrates bound at the catalytic site of EGK are shown as spacefilled models. The figure was constructed by using Deep View/Swiss-PdbViewer version 3.7 [60] and POV-Ray version 3.1 (www.povray.org) with pdb files 1glc and 1bo5.
Crystal structures of EGK do not reveal how binding of IIAGlc far from the catalytic site cleft results in allosteric inhibition. Comparison of the structures of EGK and the EGK-IIAGlc complex shows that the IIAGlc-binding site changes conformation but there are no other differences to show whether or how the catalytic site closure is affected [42]. An approach that is based on chimeric enzymes was used to identify an additional region of EGK that has a role in IIAGlc inhibition [19]. This region consists of EGK amino acid residues 427-429 and is necessary, in addition to the IIAGlc-binding site amino acid residues, to transplant IIAGlc inhibition into IIAGlc-naïve HGK. These three amino acid residues are located in the conserved ATPase core of subdomain IIA (figure 1), and this location was termed the coupling locus because it appears to be necessary to couple the IIAGlc-binding site to the catalytic site. The coupling locus amino acids affect the affinity for IIAGlc binding in the saturating presence of MgATP [19] and they modulate the local backbone motions of the IIAGlc-binding site [20]. However, the roles of the coupling locus in the allosteric coupling between the IIAGlc-binding site and the catalytic site are not known.
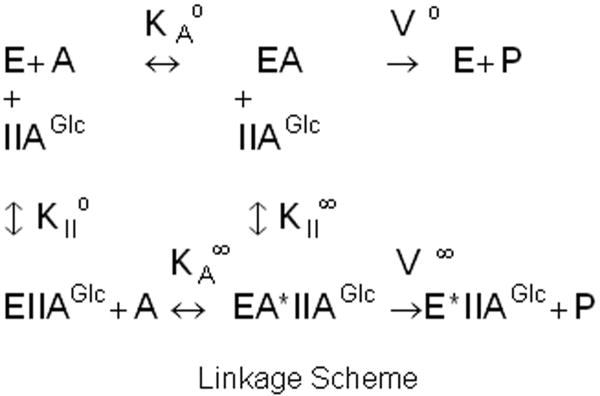
This allosteric coupling is described in terms of the thermodynamic linkage scheme that is shown in figure 2, which indicates relations between binding of IIAGlc, the Michaelis constant for MgATP, and Vmax in the context of the single-substrate, single-modifier case [43] based on the assumption of rapid equilibrium for binding of substrate [44]. The allosteric coupling is given by the parameters Q and W, which describe, respectively, the effect of binding of one ligand on the affinity for the other and the effect of the allosteric ligand on Vmax. These allosteric coupling parameters are defined by the relations: Q = KAo/KA∞ = KIIo/KII∞; W = V∞/Vo. The value of Q is >1 for cooperative coupling, <1 for antagonistic coupling, and equals 1 when there is no allosteric coupling. The value of W is >1 if Vmax is increased by the allosteric effector, <1 if Vmax is decreased by the allosteric effector, and equals 1 when there is no allosteric effect on Vmax.
Figure 2.
Thermodynamic linkage scheme for allosteric coupling between IIAGlc and MgATP. E is the EGK-glycerol complex, A is MgATP, KAo is the Michaelis constant for MgATP in the absence of IIAGlc, KA∞ is the Michaelis constant for MgATP in the saturating presence of IIAGlc, KIIo is the dissociation constant for IIAGlc binding in the absence of MgATP, KII∞ is the dissociation constant for IIAGlc binding in the saturating presence of MgATP, Vo is Vmax in the absence of IIAGlc, and V∞ is Vmax in the saturating presence of IIAGlc.
The coupling locus was identified as part of the network for allosteric inhibition because it is required for transplanting IIAGlc inhibition into HGK [19]. Substitution of only the IIAGlc-binding site amino acids from EGK into HGK to give the chimeric enzyme HGKII is not sufficient for IIAGlc binding or inhibition, but IIAGlc binding and inhibition are obtained by additional substitutions with EGK amino acids at the coupling locus. The highest affinity for IIAGlc binding to a chimeric HGK enzyme requires substitution with all three of the EGK coupling locus amino acids to give the chimeric enzyme HGKII-GTR. The native coupling locus sequence for HGK is DVN, and replacement of all three of the EGK coupling locus amino acids with those from HGK to give the chimeric enzyme EGK-DVN abolishes IIAGlc inhibition [19]. The single substitution D427G in HGKII to give the chimeric enzyme HGKII-G is sufficient for IIAGlc inhibition. However, the affinity of HGKII-G for IIAGlc is reduced greatly relative to that for HGKII-GTR, which shows that the two coupling locus amino acid residues (TR) increase the affinity for IIAGlc relative to the two native HGK amino acid residues (VN). Comparison of the allosteric coupling parameters for the chimeric enzymes HGKII-GTR and HGKII-G should show the roles of these two coupling locus amino acid residues in this allosteric network, but the IIAGlc-binding affinity of the HGKII-G variant is too low for the linked-function initial-velocity approach. However, the IIAGlc-binding affinities are sufficiently high for the corresponding EGK enzymes, which are used here to determine the role of the coupling locus in allosteric coupling. A thermodynamic linked-functions initial-velocity enzyme kinetics approach is used to determine the role of the coupling locus in the allosteric coupling between IIAGlc and MgATP, the common substrate for superfamily members. The allosteric coupling parameters are not sensitive to substitutions of the coupling locus amino acids, which indicates that this network is robust or the coupling locus is not an element of it.
Materials and Methods
Materials
Reagents were purchased from Sigma Chemical Company (St. Louis, MO) unless otherwise indicated. IIAGlc was prepared as described [45] from the BL21(DE3) strain of E. coli bearing the pVEX-crr plasmid which was generously provided by Drs. Norman Meadow and Saul Roseman of the Department of Biology of The Johns Hopkins University, Baltimore, MD. The concentration of IIAGlc was determined from absorbance at 260 nm by using an extinction coefficient of 1.6 mM-1cm-1, which was determined by using amino acid analysis to measure the molar concentration of protein in a sample for which the absorbance at 260 nm was known. Amino acid analysis was provided by the Protein Chemistry Laboratory of Texas A&M University. The molar concentration of IIAGlc was determined on the basis of a molecular mass of 18.1 kDa [46].
EGKC was prepared as described [20, 47]. EGKC-VN was constructed by using the Quick Change protocol with Pfu Turbo DNA polymerase (Stratagene; La Jolla, CA) to make the site-directed amino acid substitutions T428V and R429N in the EGKC enzyme. Mutagenic primers were purchased from Integrated DNA Technologies (Coralville, IA). The entire reading frame for the variant enzyme was verified by using the Big Dye DNA sequencing protocol. Electrophoresis of DNA sequencing reactions and Sequencher software for sequence analysis were provided by the Gene Technologies Laboratory of Texas A&M University. The EGKC-VN enzyme was purified by using the same protocol as for the EGKC enzyme. Glycerol kinase concentrations were determined from the absorbance at 280 nm by using an extinction coefficient of 1.73 (mg/mL)-1cm-1 or 97.2 mM-1(subunits)cm-1 [47].
Enzyme kinetics
Glycerol kinase enzyme activity was measured by using the continuous ADP-coupled spectrophotometric assay at pH 7.00 at 25°C to determine initial velocities. The assays contained 50 mM triethanolamine-HCl buffer, 10 mM MgCl2, 20 mM KCl, 2.5 units each of pyruvate kinase and lactate dehydrogenase, 0.2 mM PEP, 0.2 mM NADH, and 10 mM glycerol with glycerol kinase, ATP, and IIAGlc concentrations as shown in the figures in a total volume of 0.5 mL. The pH of a 2X buffer stock containing 100 mM triethanolamine-HCl, 20 mM MgCl2, and 40 mM KCl was adjusted to pH 7.00 at 25°C in a water-jacketed vessel that was connected to a circulating water bath. The temperature of the continuously-stirred solution was measured by using a digital thermometer with a small temperature probe (YSI model 402) that was standardized versus a National Bureau of Standards mercury thermometer. The pH meter was standardized to pH 7.01 at ambient temperature by using pH 7.00±0.01 commercial pH standard and the internal temperature compensator was used for measurements of solution pH at the temperature of the experiment. This 2X stock was used to prepare enzyme activity assays. Reactions were initiated by addition of enzyme (usually 0.01 mL) to the assay mixture after incubation of the remaining components in the temperature-controlled cell positioner to thermal equilibrium. The temperature of the spectrophotometer cell positioner was controlled by a refrigerated, circulating water bath which was adjusted to maintain the temperature of the experiment as determined by using the digital thermometer with the probe placed inside a water-filled cuvette. The reaction rates were determined from the rates of decrease of absorbance at 340 nm by using a Beckman-Coulter DU-800 spectrophotometer with a kinetics software package. For each assay, the observed rate was corrected by subtraction of any rate that was measured before addition of glycerol kinase. This correction is necessary at higher concentrations of IIAGlc because of an apparent ATPase activity that is associated with IIAGlc. One unit of glycerol kinase is defined as the amount of the enzyme that produces 1 μmol of ADP per minute per mL in this assay.
The stated concentrations of ATP are the total concentration of ATP in all forms. ATP concentrations were determined from absorbance at 259 nm by using an extinction coefficient of 15.4 mM-1 cm-1 for stock solutions adjusted to pH 7.0. Dilutions were performed by weight for absorbance measurements and for preparation of stock solutions in milliQ water. Dilutions were stored at -20°C.
Kinetics Data Analysis
The parameters of the linkage scheme that is shown in figure 2 are determined by using initial-velocity enzyme kinetics studies varying the concentration of ATP at fixed concentrations of IIAGlc. To obtain the parameters, the data from the initial-velocity studies are fit to equation 1 [48]
| (1) |
by using Enzfitter version 2.0.18.0 (Biosoft) running under Windows XP on a Pentium® 4 processor. The parameter uncertainties are given as the 95% confidence intervals from the fit.
Data from titrations of specific activity with IIAGlc at 2.5 mM ATP were fit to equation 2 by using Kaleidagraph v. 3.51 (Synergy Software):
| (2) |
where SAi is the specific activity at the concentration of IIAGlc given by the subscript (0 or saturating) and K0.5 is equal to the concentration of IIAGlc that gives 50% of the maximum inhibition. The allosteric coupling parameter W is given by SA∞/SA0. The parameter uncertainties are shown as standard errors.
Results
The enzymes that are used here have the substitution E478C in the IIAGlc-binding site, which is denoted by the letter C in the enzyme name. The E478C substitution increases the affinity for IIAGlc, which facilitates these determinations of the allosteric coupling. The concentration of IIAGlc that gives half-maximal inhibition of EGK is about 10 μM, which is decreased to about 1 μM by the addition of ZnCl2 or by the amino acid substitution E478C [47]. The apparent affinity is increased by Zn(II) because it binds to an intermolecular site that is formed by the EGK-IIAGlc complex [47, 49]. The protein ligands to Zn(II) are H75 and H90 from IIAGlc and E478 from EGK. While the higher affinity that is seen in the presence of Zn(II) would facilitate these studies, addition of Zn(II) complicates the analysis due to strong interactions between it and ATP, which add another simultaneous equilibrium that must be considered [47]. To avoid these issues but retain the advantages from the higher affinity, the E478C variant of each enzyme is used.2 The E478C substitution does not change the structure of the complex of EGK with IIAGlc, except for the side chain differences at the substitution site, and it eliminates the effect of Zn(II) on the apparent affinity for IIAGlc with only small effects on steady-state kinetics parameters [47].
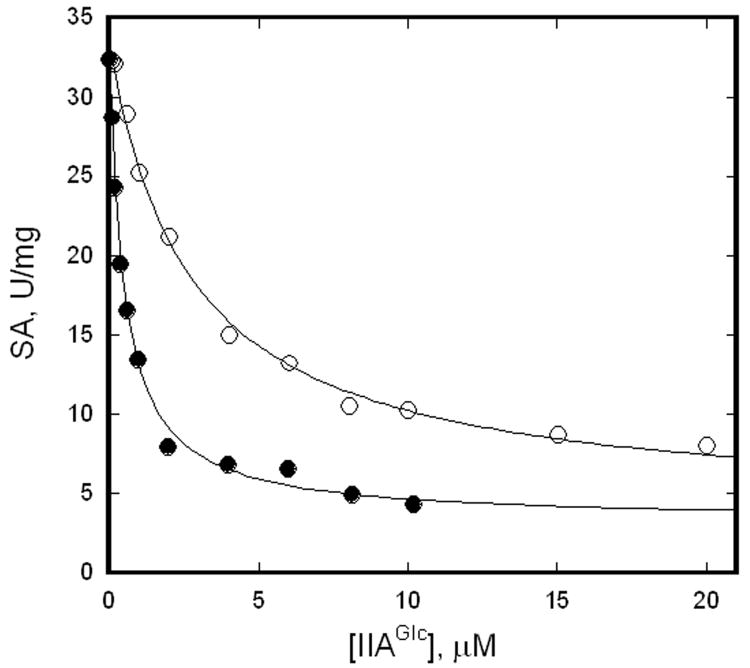
The roles of the coupling locus in IIAGlc inhibition of EGK are evaluated by comparing the allosteric properties of the enzymes EGKC and EGKC-VN. The EGKC enzyme contains the EGK native coupling locus, while the EGKC-VN enzyme contains site-directed substitutions T428V and R429N to replace two coupling locus amino acids with those from HGK. Thus, EGKC-VN corresponds to HGKII-G, the minimal IIAGlc-inhibitable HGK, while EGKC corresponds to HGKII-GTR. Figure 3 shows IIAGlc inhibition of these enzymes at saturating concentrations of glycerol and MgATP. The Michaelis constant for glycerol is about 5 μM for both EGKC and EGK, and IIAGlc decreases it for EGK [47]. Thus, the glycerol concentration of 10 mM is saturating. Saturation with MgATP is verified by results that are shown below. The specific activity decreases as the concentration of IIAGlc is increased for both enzymes. These data are fit to equation 2 to obtain estimates of K0.5 and W, which are given in the figure legend.
Figure 3.
IIAGlc inhibition of EGKC and EGKC-VN. Specific activities were measured at 9 nM (subunits) enzyme with 2.5 mM ATP and IIAGlc concentrations as shown. The points show the specific activities and the lines show the fits to equation 2 for the parameters given below. Legend: filled circles, EGKC, K0.5 = 0.5±0.1 μM, W = 0.1±0.01; open circles, EGKC-VN, K0.5 = 2.7±0.3 μM, W = 0.11±0.02.
The inhibition shows hyperbolic dependence on IIAGlc concentration and is well- described by equation 2. The parameters K0.5 and W that are determined for EGKC agree with results from earlier work: K0.5 = 0.6±0.2 μM, W = 0.12±0.04 [47]. Thus, the same parameters are obtained with different protein preparations and over a long time span. The coupling locus substitutions in EGKC-VN reduce apparent affinity for IIAGlc by about 5-fold. The reduced affinity is consistent with the relation between the coupling locus and IIAGlc affinity that is seen in the studies for transplantation of IIAGlc inhibition into HGK [19]. The same value for the allosteric coupling parameter W is obtained for EGKC and EGKC-VN, which shows that it is not changed by the coupling locus substitutions. The linkage scheme in figure 2 shows that K0.5 that is determined from these data corresponds to KII∞, the dissociation constant for IIAGlc binding in the saturating presence of MgATP. These results that are obtained with saturating MgATP reveal the effect of IIAGlc binding on Vmax but not the allosteric coupling between the binding of MgATP and IIAGlc.
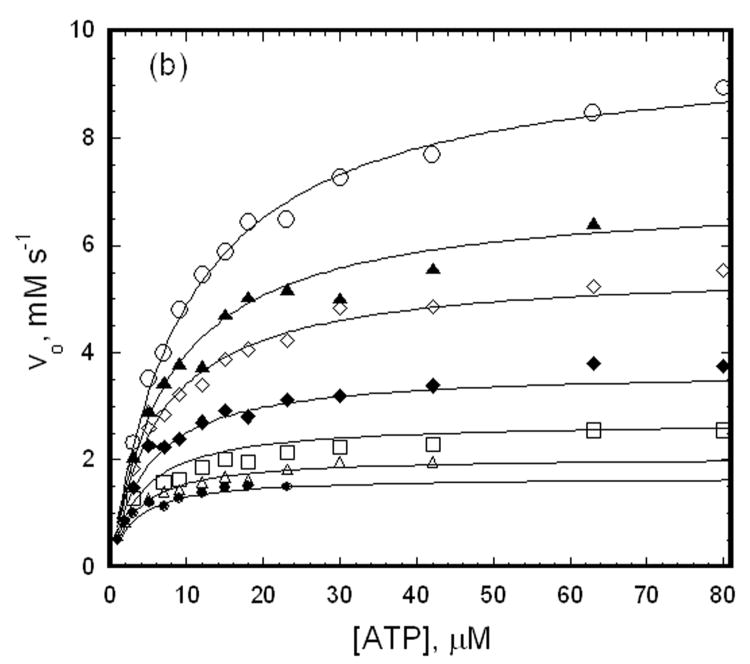
The approach that is used here to determine the effects of the coupling locus substitutions on allosteric coupling between binding of MgATP and IIAGlc is linked-functions initial-velocity enzyme kinetics studies that are treated in the context of the linkage scheme that is shown in figure 2. Although the apparent affinity for IIAGlc is reduced by the amino acid substitutions in EGKC-VN, it is sufficiently high for this linked-functions analysis, which is not the case for the chimeric HGK enzymes from the IIAGlc-inhibition transplantation studies. Figure 4 shows the dependence of the rate of the enzymatic reaction on the concentration of ATP in the saturating presence of glycerol with different fixed levels of IIAGlc for EGKC and EGKC-VN. As the concentration of IIAGlc is increased from zero, the initial velocities are decreased throughout the range of ATP concentrations. These data are fit to equation 1 to obtain the estimates of the initial-velocity kinetics parameters and allosteric coupling parameters that are shown in Table 1. Fits of simulated data with varying levels of random error were used to show that the parameter estimation procedure is very reliable (see Supplementary Information).
Figure 4.
Effects of IIAGlc on initial velocities for EGKC and EGKC-VN. The points show initial velocities that are measured at 27 nM (subunits) enzyme and the ATP concentrations that are shown. The open circles show the initial velocities in the absence of IIAGlc, and the remaining symbols show the initial velocities when IIAGlc is added to the assay at the concentrations (μM) indicated below, increasing from the upper curve to the lower curve of each figure. The lines show the fits of the data to equation 1 yielding the parameters that are shown in Table 1. (a) EGKC, IIAGlc: 0.11, 0.26, 0.52, 1.0, 2.0, 5.2, 10. (b) EGKC-VN, IIAGlc: 1, 2, 5.2, 10, 20, 40.
Table 1.
Effects of the coupling locus changes on catalytic and allosteric coupling parametersa
| Enzyme | Vo, mM s-1 | KAo, μM | KIIo, μM | Q | W | KII∞, μM |
|---|---|---|---|---|---|---|
| EGKC | 8 (7, 9) | 9 (8, 10) | 1.3 (0.8, 1.9) | 2 (1, 3) | 0.12 (0.09, 0.14) | 0.6 (0.25, 1.1) |
| EGKC-VN | 9.5 (9, 10) | 10 (9, 11) | 8 (5, 11) | 4 (2, 6) | 0.13 (0.11, 0.15) | 2.1 (0.8, 3.4) |
The parameter values are estimated from simultaneous fits of the data in figure 4 to equation 1 except for KII∞ which is given by the definition of the allosteric coupling parameter Q, KII∞ = KIIo/Q. The 95% confidence intervals for the parameter estimates from the fit are shown in parenthesis, and the interval for KII∞ is propagated from those for KIIo and Q.
The values of the parameters are well-determined as shown by the 95% confidence intervals and by the agreement between the data and the lines from the fits in figure 4. Neither the catalytic parameters Vo and KAo nor the allosteric coupling constants Q and W are changed significantly by the coupling locus substitutions. The Michaelis constant for MgATP for each enzyme, KAo, is 10 μM, showing that the ATP concentration of 2.5 mM that is used for the experiment in figure 3 is saturating. The allosteric coupling constant Q is about 3 for both enzymes, showing weak coupling between binding of MgATP and IIAGlc. The dominant effect of IIAGlc on enzymatic activity is thus reduction of Vmax, i.e. V-type control, shown by the allosteric coupling parameter W of 0.1. The coupling locus substitutions significantly change the dissociation constant for IIAGlc binding to the GK-glycerol complex, KIIo, which is increased about 5-fold for EGKC-VN relative to EGKC.
The linked-functions relations that are given by the linkage scheme can be used to evaluate the internal consistency of the estimated parameters. The linkage scheme in figure 2 shows KII∞ is the dissociation constant for binding of IIAGlc to the enzyme-glycerol-MgATP complex. The constant K0.5 from figure 3 is determined at saturating MgATP; thus, it corresponds to KII∞, leading to the expectation that the values of these constants that are obtained from independent experiments should agree. The definition of the allosteric coupling constant Q gives KII∞ as KIIo/Q, thus allowing its calculation to give the values that are shown in Table 1. They agree with the values of K0.5 that are shown in figure 3. For both enzymes, the values for the allosteric coupling parameter W are the same irrespective of whether ATP or IIAGlc is the varied ligand. The analyses of the allosteric coupling between MgATP and IIAGlc are thus internally consistent with respect to both coupling parameters. The values for KIIo and K0.5 are changed about 5-fold by the substitutions, showing that the value of the allosteric coupling constant Q is not changed.
The allosteric coupling constant Q is greater than 1 for both enzymes, showing that the coupling between MgATP and IIAGlc is cooperative. In the context of the scheme in figure 2, the values of Q show that KII∞<KIIo and KA∞<KAo. These ligands thus mutually increase the apparent affinity for each other. Cooperative coupling between substrates and allosteric ligands is associated with activation for classically-studied K-type allosteric enzymes [50-54] but is here associated with inhibition of catalysis. If the allosteric coupling between IIAGlc and the reaction products is also cooperative, increased affinity for the products could account for a portion of the reduction in Vmax by IIAGlc. The coupling free energy between MgATP and IIAGlc binding is small; its value given by –RTlnQ is about -0.6 kcal/mol.
The allosteric coupling constant W for both enzymes shows that V∞<Vo; hence Vmax is decreased by binding of IIAGlc, as expected for inhibition. The values for W for both enzymes are greater than zero, i.e. V∞>0, showing that inhibition is incomplete and enzyme that is saturated with IIAGlc retains about 10% catalytic activity relative to that in the absence of the inhibitory ligand. Such partial inhibition is consistent with allosteric inhibition. The increase of apparent activation energy for the reaction in the saturating presence of IIAGlc given by –RTlnW is about +1.3 kcal/mol.
Discussion
The objective of these studies is determination of the role of the coupling locus in the allosteric network that couples the allosteric and catalytic sites, for which measurement of the allosteric coupling parameters is required. The allosteric parameters that describe the coupling between binding of IIAGlc and MgATP and the effect of IIAGlc on Vmax for EGKC and EGKC-VN are determined by using a linked-functions initial-velocity enzyme kinetics approach. Results from this approach show that substitutions of the coupling locus amino acids change the affinity for IIAGlc but do not alter either of the quantitative measures of allosteric coupling between the catalytic and allosteric sites, Q and W. Thus, IIAGlc-binding affinity and MgATP-IIAGlc allosteric coupling are independent from one another. The linkage scheme in figure 2 shows no requirement for specific relations between allosteric ligand affinity and allosteric coupling. Thus, the independence of the affinity and allosteric coupling that is observed here is unremarkable in the context of a phenomenological treatment of allosteric behavior. Independence of ligand affinity and allosteric coupling is seen also for bacterial phosphofructokinase for two different allosteric inhibitors that bind to the same site [54] and for substitutions of substrate–binding amino acid residues [55].
The catalytic parameters and the allosteric coupling parameters are insensitive to the coupling locus substitutions. This result may indicate that the catalytic and allosteric networks are robust or the coupling locus is not an element of these networks. Because the coupling locus is located in the conserved ATPase core, the insensitivity of the catalytic parameters to the substitutions may reflect the robustness of the catalytic network. Such robustness is consistent with views on protein evolution, which consider that amino acid substitutions improve a new function without being deleterious to an original function [56]. This behavior is the case for improvement of the affinity for the allosteric inhibitor IIAGlc without affecting the ATPase catalytic activity. Limited structural information about conformational changes upon binding of ATP or ADP to sugar kinases shows small, localized changes in domain I but none in domain II [24, 27]. The absence of nucleotide substrate-dependent conformational changes in the region that contains the coupling locus may indicate that it is not part of the catalytic network.
The ATPase core of the superfamily member DnaK is established as a key element of its allosteric network [18]. Deuterium exchange/mass spectrometry studies show conformational changes within the core that mediate communication between the catalytic site and the peptide substrate binding site. The structure of the yeast hsp70 homologue Sse1 shows that the linker which connects the nucleotide- and substrate-binding domains binds in the cleft between domains I and II of the nucleotide-binding domain in the absence of peptides [38]. The conformational changes within the core upon peptide binding that are measured in the deuterium exchange/mass spectrometry studies are consistent with displacement of the linker from its binding site. Thus, allosteric control of the DnaK ATPase activity appears to be mediated by direct steric effects of linker binding to both domains of the conserved core. Such direct effects are not the case for IIAGlc allosteric control of EGK because the IIAGlc-binding site is far from the catalytic cleft and contained in only one domain. Because of the difference in the relations of the allosteric and catalytic sites of EGK relative to other superfamily members, structural elements outside the conserved ATPase core may be the principal mediators of the allosteric network for IIAGlc inhibition. Primary roles for these other elements in allosteric communication may account for the insensitivity of the allosteric coupling parameters to the coupling locus substitutions.
The oligomeric structure of EGK differs from that of other superfamily members and may provide for allosteric communication outside the conserved ATPase core. Other allosteric superfamily members are monomers in solution but EGK displays a dimer-tetramer equilibrium [57]. Disruption of tetramer formation abolishes allosteric inhibition by fructose 1,6-bisphosphate but has little effect on IIAGlc inhibition [58, 59]. Within the EGK dimer, a loop from one subunit penetrates deeply into the catalytic cleft of the other subunit. This interdigitated structure could have roles in the IIAGlc allosteric network providing for interdomain communication by pathways outside the conserved ATPase core. Alternatively, allosteric networks may include elements of the conserved ATPase core as well as the elements of the interdigitated structure. In this case, the more extensive networks could be expected to show substantial robustness that is seen as the insensitivity of the allosteric coupling parameters to the coupling locus substitutions.
In its initial formulation, the concept of allostery refered to change in affinity for ligand binding at one site that is a consequence of ligand binding at a different site and the changed affinity was attributed to global conformational change associated with altered protein quaternary structure [1]. Current views consider that changes in protein conformation and/or dynamics in response to ligand binding, post-translational modification, or amino acid substitutions are allosteric and may occur within monomeric, single-domain proteins. The changes in protein conformation and/or dynamics are not global but involve networks of amino acid residues that transmit allosteric signals within the protein structure. This view of allostery raises important considerations. A primary issue is identification of the elements of the allosteric network. In this context, it is important to note that the elements depend on the function that is studied so that different functions may arise from different networks. A second issue is verification that network components that are putatively identified on the basis of, for example, structural considerations are members of the functional network of interest. Verification of network membership requires application of double-mutant cycles or linked-functions approaches [8]. The importance of these two issues is supported by the results that are presented here. Although it is clear that there is an allosteric network between the coupling locus and the IIAGlc allosteric binding site, the results raise questions about whether the coupling locus is an element of a network between the IIAGlc allosteric site and the catalytic site.
Supplementary Material
Acknowledgments
This work was supported by grant GM068768 from the National Institutes of Health and by Texas AgriLife Research (formerly Texas Agricultural Experiment Station). The author thanks Jesse Flynn, Rebecca Jurrens, Pamela S. Miller, Brandon Sexton, and Jillian Wisdom for expert technical assistance; the Protein Chemistry Laboratory of Texas A&M University for performing amino acid analysis for determination of the extinction coefficient for IIAGlc; the Gene Technologies Laboratory of Texas A&M University for DNA sequencing; Drs. Norman Meadow and Saul Roseman for generously providing the E. coli strain bearing the plasmid for expression of IIAGlc and helpful discussions about the purification of IIAGlc; and Dr. Gregory D. Reinhart for helpful discussions.
Footnotes
Abbreviations used: IIAGlc the glucose-specific phosphocarrier protein of the phosphoenolpyruvate:glycose phosphotransferase system (TC 4.A.1 (http://www.tcdb.org/)), also known as IIIglc and Crr; EGK, Escherichia coli glycerol kinase (EC 2.7.1.30; ATP:glycerol 3-phosphotransferase); EGKC, EGK with the amino acid substitution E478C; EGKC-VN, EGKC with the amino acid substitutions T428V and R429N; EGK-DVN, EGK with the amino acid substitutions G427D, T428V, and R429N; HGK, Haemophilus influenzae glycerol kinase; HGKII, HGK with IIAGlc-binding site amino acids (II) from EGK; HGKII-G, HGKII with amino acid substitution D427G; HGKII-GTR, HGKII with the amino acid substitutions D427G, V428T, N429R; hsp70, heat shock protein 70.
The IIAGlc binding affinity is increased for the HGKIIC-G chimeric enzyme but is too low for the linked-function initial-velocity studies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Monod J, Wyman J, Changeaux J-P. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Rosseau F, Schymkowitz J. Curr Opin Struct Biol. 2005;15:23–30. doi: 10.1016/j.sbi.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Daily MD, Upadhyaya TJ, Gray JJ. Proteins. 2008;71:455–466. doi: 10.1002/prot.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, Whitten S, Hilser VJ. Proc Natl Acad Sci USA. 2007;104:4347–4352. doi: 10.1073/pnas.0607132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodey N, Benkovic SJ. Nature Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 7.Suel G, Lockless S, Wall M, Ranganathan R. Nature Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 8.Ackers GK, Smith FR. Annu Rev Biochem. 1985;54:597–629. doi: 10.1146/annurev.bi.54.070185.003121. [DOI] [PubMed] [Google Scholar]
- 9.del Sol A, Arauzo-Bravo MJ, Amoros D, Nussinov R. Genome Biol. 2007;8:R92. doi: 10.1186/gb-2007-8-5-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dima R, Thirumalai D. Protein Sci. 2006;15:258–268. doi: 10.1110/ps.051767306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daily MD, Gray JJ. Proteins. 2007;67:385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 12.Amaro R, Sethi A, Myers R, Davisson V, Luthey-Schulten Z. Biochemistry. 2007;46:2156–2173. doi: 10.1021/bi061708e. [DOI] [PubMed] [Google Scholar]
- 13.Luque I, Freire E. Proteins. 2000;4:63–71. doi: 10.1002/1097-0134(2000)41:4+<63::aid-prot60>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- 14.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. J Mol Biol. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Lukasik SM, Liu Y, Grembecka J, Bielnicka I, Bushweller JH, Speck NA. J Mol Biol. 2006;364:1073–1083. doi: 10.1016/j.jmb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer JA, Lee AL. Biochemistry. 2008;47:4876–4886. doi: 10.1021/bi702330t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Z, Resing KA, Ahn NG. Curr Opin Struct Biol. 2006;16:686–692. doi: 10.1016/j.sbi.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Rist W, Graf C, Bukau B, Mayer MP. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 19.Pawlyk AC, Pettigrew DW. Proc Natl Acad Sci USA. 2002;99:11115–11120. doi: 10.1073/pnas.132393599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu P, Lasagna M, Pawlyk A, Reinhart GD, Pettigrew DW. Biochemistry. 2007;46:12355–12365. doi: 10.1021/bi7010948. [DOI] [PubMed] [Google Scholar]
- 21.Hogema BM, Arents JC, Bader R, Eijkemans K, Yoshida H, Takahashi H, Alba H, Postma PW. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- 22.Postma PW, Lengeler JW, Jacobson GR. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roseman S. In: Phosphate in Microorganisms: Cellular and Molecular Biology. Torriani-Gorini A, Yagil E, Silver S, editors. ASM Press; Washington, D. C.: 1994. pp. 151–160. [Google Scholar]
- 24.Hurley JH, Faber HR, Worthylake D, Meadow ND, Roseman S, Pettigrew DW, Remington SJ. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 25.Hurley JH. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 26.Gerstein M, Lesk AM, Chothia C. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 27.Grueninger D, Schulz GE. J Mol Biol. 2006;359:787–797. doi: 10.1016/j.jmb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Aleshin AE, Kirby C, Liu X, Bourenkov GP, Bartunik HD, Fromm HJ, Honzatko RB. J Mol Biol. 2000;296:1001–1015. doi: 10.1006/jmbi.1999.3494. [DOI] [PubMed] [Google Scholar]
- 29.Graceffa P, Dominguez R. J Biol Chem. 2003;278:34172–34180. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 30.Chu J-W, Voth GA. Proc Natl Acad Sci USA. 2005;102:13111–13116. doi: 10.1073/pnas.0503732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page R, Lindberg U, Schutt CE. J Mol Biol. 1998;280:463–474. doi: 10.1006/jmbi.1998.1879. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zuiderweg ERP. Proc Natl Acad Sci USA. 2004;101:10272–10277. doi: 10.1073/pnas.0401313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh JI, Charrier V, Paulo J, Hou L, Darbon E, Claiborne A, Hol WGJ, Deutscher J. Biochemistry. 2004;43:362–373. doi: 10.1021/bi034258o. [DOI] [PubMed] [Google Scholar]
- 34.Kamata K, Mitsuya M, Nishimura T, Eiki J-i, Nagata Y. Structure. 2004;12:429–438. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Otterbein LR, Cosio C, Graceffa P, Dominguez R. Proc Natl Acad Sci USA. 2002;99:8003–8008. doi: 10.1073/pnas.122126299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominguez R. TIBS. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Hendrickson WA. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F-U, Kuriyan J. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 40.Mayer MP, Bukau B. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sondermann H, Scheufler C, Schneider C, Hohfled J, Hartl F-U, Moarefi I. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 42.Feese MD, Faber HR, Bystrom CE, Pettigrew DW, Remington SJ. Structure. 1998;6:1407–1418. doi: 10.1016/s0969-2126(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 43.Frieden C. J Biol Chem. 1964;239:3522–3531. [PubMed] [Google Scholar]
- 44.Reinhart GD. In: Energetics of Biological Macromolecules, Part E. Holt Jo M, J ML, Ackers Gary K., editors. Academic Press; 2004. pp. 187–203. [Google Scholar]
- 45.Pelton JG, Torchia DA, Meadow ND, Wong C-Y, Roseman S. Biochemistry. 1991;30:10043–10057. doi: 10.1021/bi00105a032. [DOI] [PubMed] [Google Scholar]
- 46.Worthylake D, Meadow N, Roseman S, Liao D-I, Herzberg O, Remington SJ. Proc Natl Acad Sci USA. 1991;88:10382–10386. doi: 10.1073/pnas.88.23.10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettigrew DW, Meadow ND, Roseman S, Remington SJ. Biochemistry. 1998;37:4875–4883. doi: 10.1021/bi971634u. [DOI] [PubMed] [Google Scholar]
- 48.Reinhart GD. Arch Biochem Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 49.Feese M, Pettigrew DW, Meadow ND, Roseman S, Remington SJ. Proc Natl Acad Sci USA. 1994;91:3544–3548. doi: 10.1073/pnas.91.9.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mesecar AD, Nowak T. Biochemistry. 1997;36:6792–6802. doi: 10.1021/bi962869t. [DOI] [PubMed] [Google Scholar]
- 51.Johnson JL, Reinhart GD. Biochemistry. 1992;31:11510–11518. doi: 10.1021/bi00161a032. [DOI] [PubMed] [Google Scholar]
- 52.Symcox MM, Reinhart GD. Analytical Biochem. 1992;206:394–399. doi: 10.1016/0003-2697(92)90384-j. [DOI] [PubMed] [Google Scholar]
- 53.Braxton BL, Tlapak-Simmons VL, Reinhart GD. J Biol Chem. 1994;269:47–50. [PubMed] [Google Scholar]
- 54.Tlapak-Simmons VL, Reinhart GD. Arch Biochem Biophys. 1994;308:226–230. doi: 10.1006/abbi.1994.1032. [DOI] [PubMed] [Google Scholar]
- 55.Fenton AW, Paricharttanakul NM, Reinhart GD. Biochemistry. 2003;42:6453–6459. doi: 10.1021/bi034273t. [DOI] [PubMed] [Google Scholar]
- 56.Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, Tawfik DS. Nature Genetics. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 57.de Riel JK, Paulus H. Biochemistry. 1978;17:5141–5146. doi: 10.1021/bi00617a011. [DOI] [PubMed] [Google Scholar]
- 58.Liu WZ, Faber R, Feese M, Remington SJ, Pettigrew DW. Biochemistry. 1994;33:10120–10126. doi: 10.1021/bi00199a040. [DOI] [PubMed] [Google Scholar]
- 59.Bystrom CE, Pettigrew DW, Branchaud BP, O’Brien P, Remington SJ. Biochemistry. 1999;38:3508–3518. doi: 10.1021/bi982460z. [DOI] [PubMed] [Google Scholar]
- 60.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.