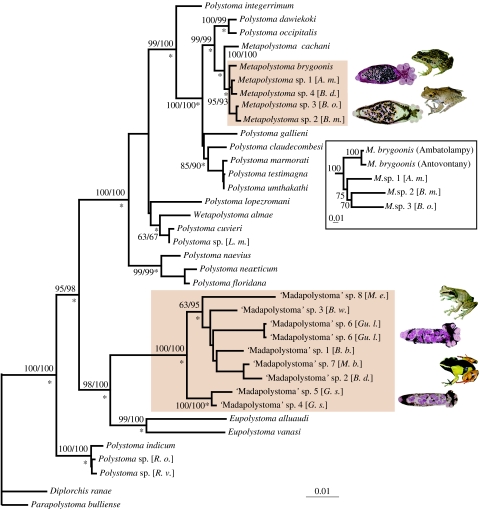
Figure 1.
Best maximum likelihood (ML) tree inferred from the analysis of complete 18S and partial 28S sequences. Abbreviations in brackets refer to host species, from top to bottom: B. m., Boophis madagascariensis; B. o., Boophis occidentalis; A. m., Aglyptodactylus madagascariensis; B. d., Boophis doulioti; L. m., Leptodactylus mystaceus; M. e., Mantella expectata; B. w., Blommersia wittei; Gu. l., Guibemantis liber; B. b., Blommersia blommersae; M. b., Mantella baroni; B. d., Blommersia domerguei; G. s., Gephyromantis sculpturatus; R. o., Rhacophorus omeimontis; R. v., Rhacophorus viridis. Values above or below branches indicate, respectively, maximum parsimony (MP) and ML bootstrap proportions (BPs) after 1000 replicates and asterisks refer to Bayesian posterior probabilities (BPPs) superior to 0.95. The two Malagasy polystome radiations are in brown boxes. Inset photos show M. brygoonis and M. sp. 2 with their host species, P. mascareniensis and Boophis madagascariensis; and two undescribed species of ‘Madapolystoma’ with their host species, Mantella madagascariensis (not included in this analysis, but one of the few host species that yielded a mature ‘Madapolystoma’) and G. liber. The inset tree shows relationships among some Malagasy Metapolystoma based on partial COI sequences, indicating the distinct genetic differentiation and putative basal position of M. brygoonis from the host species P. mascareniensis relative to the specimens recovered from mantellid hosts.