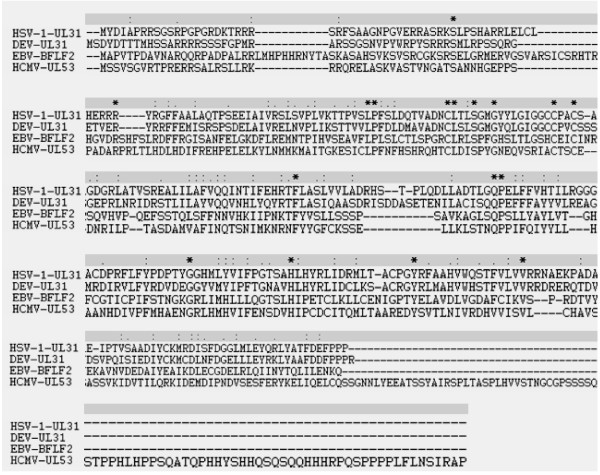
Figure 2.
Amino acid sequence comparison between the putative proteins encoded by DEV UL31 and it homologs in hunman herpesviruses: HSV-1 UL31, HCMV UL53, and EBV BFLF2. Sequences were aligned with the Clustalx1.8 software. Absence of amino acid is shown by dash '-' in the sequences while '*', ':', and '.' Indicate identical amino acid residues, conserved residues and semi-conserved residues in all sequence used in the alignment respectively.