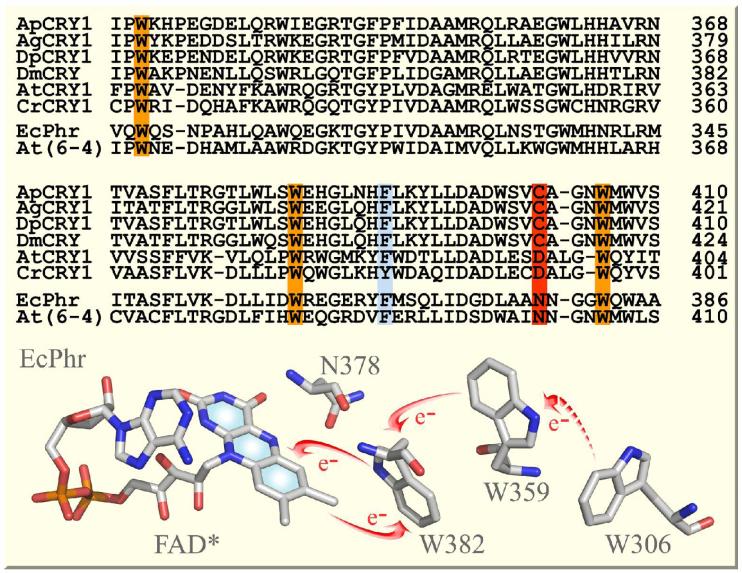
Figure 2.
Sequence alignment of four insect Type 1 CRYs, two plant CRYs, and two photolyases. Ap, Antheraea pernyi (Chinese oak silk moth); Ag, Anopheles gambiae (mosquito); Dp, Danaus plexippus (monarch butterfly); Dm, Drosophila melanogaster (fruit fly); At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Ec, Escherichia coli. Note the conserved tryptophan triad (in orange) for photoreduction through electron transfer across all CRYs and photolyases. Another nearby aromatic residue F or Y (in blue) may also make contributions. Note the critical conserved residue C in insect Type 1 CRYs, N in photolyases and D in plant CRYs (in red) near the N5 position of the isoalloxazine ring, which determines the protonation state of the reduced flavin. Shown at the bottom is the local X-ray structure around the cofactor in E. coli photolyase20 with arrows indicating electron transfer pathways.