Figure 4.
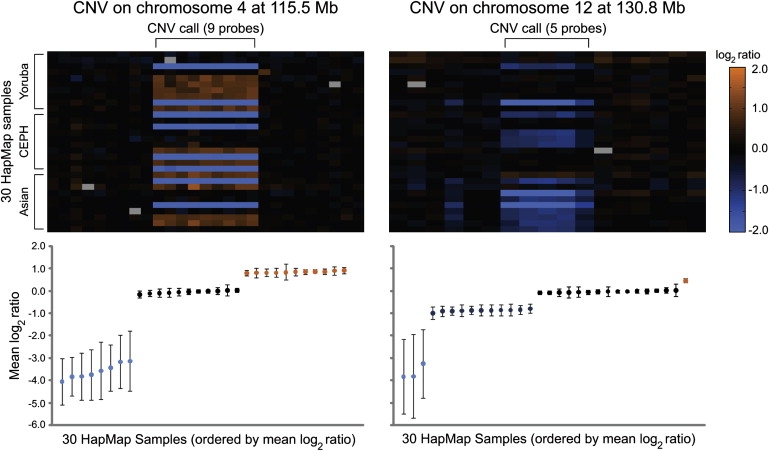
Simple CNVs and Inference of Genotypes, Based on Discrete Log2-Ratio Clustering
For two CNV-containing genomic regions that have similar estimated breakpoints across all individuals, probe-by-probe log2 ratios are depicted in heatmaps (see scale bar) in the upper panel (with rows representing individuals and columns representing probes ordered by genomic position). Mean log2 ratios of the probes within the CNV are provided in the lower panel. The mean log2 ratios form discrete clusters, letting us infer CNV genotypes. For both loci, there is one cluster with strongly negative log2 ratios, suggesting that these individuals have homozygous deletions for this DNA segment. For the CNV on chromosome 4 at 155.5 Mb (hg17), there are three mean log2-ratio clusters, likely reflecting zero, one, and two copies of this DNA segment. For the CNV on chromosome 12 at 130.8 Mb there are four mean log2-ratio clusters, likely reflecting states of zero, one, two, and three copies; therefore, this CNV would be considered to be multiallelic. Error bars represent the standard deviation (SD).
