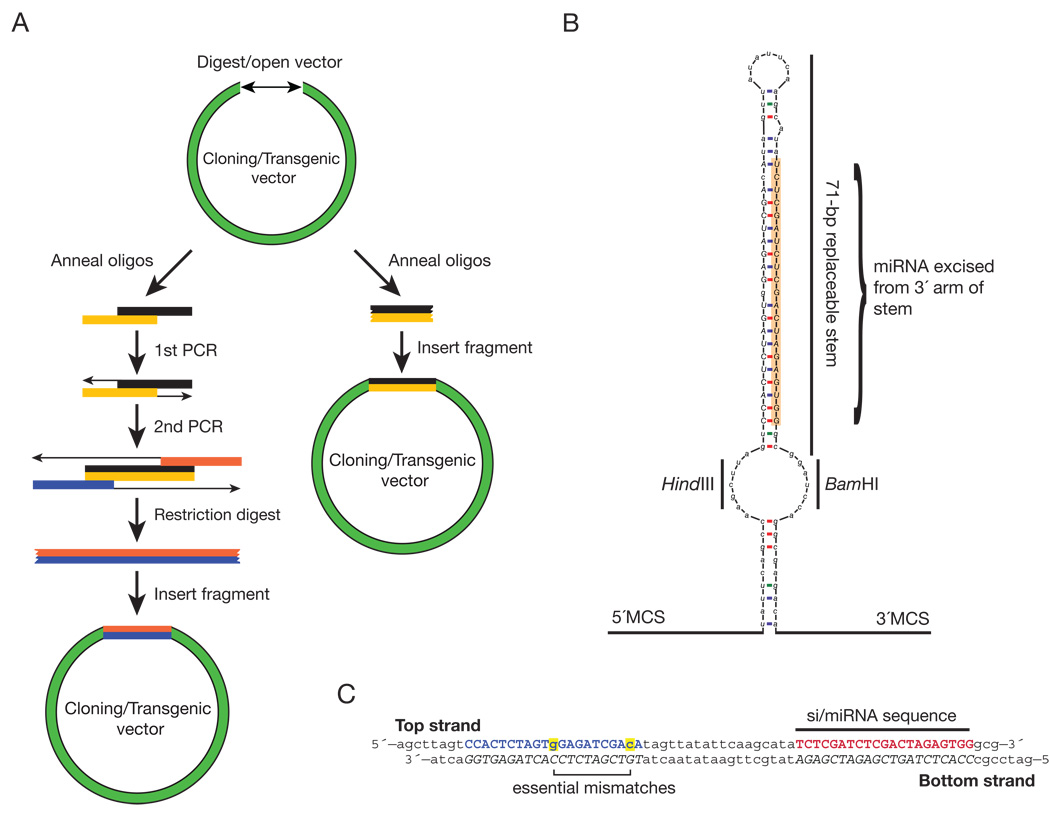
Fig. 1. Schematic of miRNA-based RNAi vector—shmiRs.
(A) Standard method for cloning miRNA-based RNAi vectors, left, is compared to miRNA-based vectors described in this study, right. (B) Structure of shmiR-dpp2 in pHB scaffold. Annealed 71-nt DNA oligos are inserted into the open-loop region with the specified restriction sequences, HindIII-BamHI in this example. Resultant siRNAs will be excised, like miR-1, from the 3′ arm of the stem-loop (orange highlighted region). This construct is flanked by 5′ and 3′ cloning sites (M.C.S.), enabling the creation of polycistronic hairpins. (C) shmiR-dpp2 knockdown oligos inserted into the HindIII-BamHI site of pHB. The siRNA sequence is highlighted in red, while the blue nucleotides represent the presumptive star sequence of the dpp2 siRNA. Highlighted in yellow are two specific mismatched bases necessary to maintain the endogenous pre-miR-1 stem-loop structure.