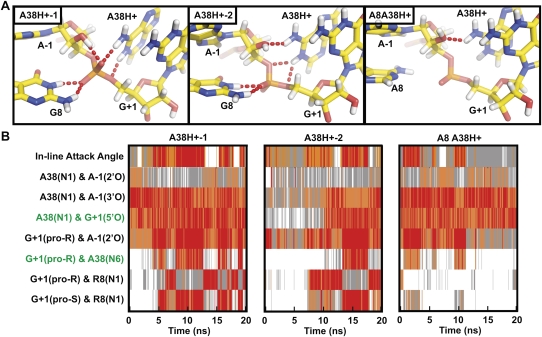
FIGURE 6.
Protonation of A38(N1) results in a conformational rearrangement around the active site that establishes structural features predicted to be present within the transition state. (A) Snapshots of the active site from three simulations (averaged either from 9.9 to 10 ns for A38H+-1 or from 19.9 to 20 ns for A38H+-2 and A8A38H+) in which N1 of A38 is protonated. Key hydrogen bonds are indicated by red dash tubes. (B) In-line attack angle and important interatomic distances are tracked over the course of the simulation, with distances and angles color coded as in Figure 4. Proximity of the cleavage site G+1(5′O) and A38(N1) is frequently observed in all three simulations. A favorable IAA coincides with rotation of the nonbridging oxygens such that of G8(N1) loses its hydrogen bonding with the pro-R nonbridging oxygen in favor of a hydrogen bond with the pro-S nonbridging oxygen, as illustrated in the A38H+-1 snapshot of (A). Hydrogen bonding partners observed in TSA crystal structures (2P7E, 2P7F, and 1M50, indicated in green) are more frequently observed in these simulations.