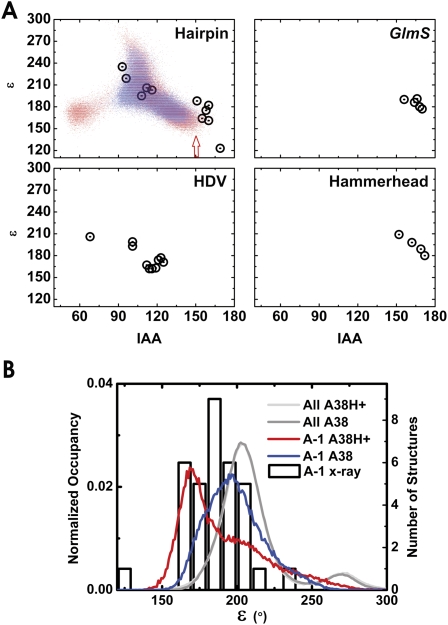
FIGURE 7.
The cleavage site dynamics of the RNA backbone of our simulations share conformational features common among the small self-cleaving RNAs. (A) Cleavage site dihedral angle epsilon as a function of the IAA for all crystal structures with <3.1 Å resolution of the hairpin ribozyme, the glmS ribozyme bound to glucose amine 6-phosphate, the HDV ribozyme, and the full-length hammerhead ribozyme (open circles). Epsilon as a function of IAA is also shown for the A38H+ simulations (red dots), as well as the C39-1 and C39-2 simulations (blue dots). The A38H+ simulations sample a wider range of values in these plots; notably, the region of both lowest epsilon and highest IAA (red arrow) is more frequently sampled in the A38H+ simulations. (B) Probability densities of the cleavage site (A-1) epsilon angles as well as all backbone epsilon angles in our simulations (left axis and lines) and cleavage site (A-1) epsilon distributions among the available ribozyme crystal structures (right axis and open boxes). The epsilon angles at the cleavage site during our simulations and in the crystal structures both favor low values relative to the distribution observed in our simulations for all epsilon angles in the entire backbone. Moreover, simulations in which A38 is protonated (red line) favor even lower epsilon values than simulations where A38 is unprotonated (blue line).