Abstract
Goodpasture-antigen binding protein (GPBP) is a nonconventional Ser/Thr kinase for basement membrane type IV collagen. Various studies have questioned these findings and proposed that GPBP serves as transporter of ceramide between the endoplasmic reticulum and the Golgi apparatus. Here we show that cells expressed at least two GPBP isoforms resulting from canonical (77-kDa) and noncanonical (91-kDa) mRNA translation initiation. The 77-kDa polypeptide interacted with type IV collagen and localized as a soluble form in the extracellular compartment. The 91-kDa polypeptide and its derived 120-kDa polypeptide associated with cellular membranes and regulated the extracellular levels of the 77-kDa polypeptide. A short motif containing two phenylalanines in an acidic tract and the 26-residue Ser-rich region were required for efficient 77-kDa polypeptide secretion. Removal of the 26-residue Ser-rich region by alternative exon splicing rendered the protein cytosolic and sensitive to the reduction of sphingomyelin cellular levels. These and previous data implicate GPBPs in a multicompartmental program for protein secretion (i.e. type IV collagen) that includes: 1) phosphorylation and regulation of protein molecular/supramolecular organization and 2) interorganelle ceramide trafficking and regulation of protein cargo transport to the plasma membrane.
Goodpasture antigen-binding protein (GPBP)2 phosphorylates the noncollagenous-1 (NC1) domain of the α3 chain of type IV collagen (α3(IV)NC1) (1). This domain is a pivotal structure in the molecular and supramolecular organization of the glomerular basement membrane collagen and also the target of autoantibodies mediating glomerulonephritis in Goodpasture disease (2). Increased GPBP expression has been associated with autoimmune pathogenesis including Goodpasture disease (3) and with the induction of glomerular basement membrane collagen disorganization and deposit of IgA antibodies (4). These observations suggest that GPBP regulates glomerular basement membrane collagen organization and induces type IV collagen-based antibody-mediated glomerulonephritis when its expression is abnormally elevated (3, 4). The GPBP gene (COL4A3BP) also encodes for GPBPΔ26, a more abundant, less active, alternatively spliced GPBP variant lacking a 26-residue Ser-rich region that is apparently not regulated under these pathological conditions (3).
GPBP contains multiple structural elements including N-terminal pleckstrin homology domain, Ser-Xaa-Yaa region, bipartite nuclear localization signal, coiled-coil domain, two phenylalanines in an acidic tract (FFAT) motif, and C-terminal steroidogenic acute regulatory related lipid transfer (START) domain. Additional structural features include motifs for self-interaction and phosphorylation (1, 3, 5, 6). The pleckstrin homology domains comprise a variety of poorly conserved structures present only in eukaryotes that have been proposed to mediate protein targeting to cellular membranes through interaction with phosphoinositides (7). A variety of proteins including several protein kinases contain pleckstrin homology domains (8). The FFAT motifs target proteins to the endoplasmic reticulum (ER) through interaction with the transmembrane cytosolic domain of the vesicle-associated membrane protein-associated proteins (VAPs) (9), which have been proposed to play a role in maintaining homeostasis for protein folding in the ER and in regulating protein cargo transport to the plasma membrane (10, 11). The START domains bind lipids including ceramide, phospholipids, and sterols and are modules present in a variety of proteins with distinct physiological and pathological functions (12, 13).
Recent reports have implicated the FFAT motif and pleckstrin homology domain in the binding of GPBP polypeptides to the ER and Golgi apparatus, respectively. The binding to these organelles has been postulated to enable the START domain to capture ceramide from the ER and to deliver it to the Golgi apparatus. Based on these observations, GPBP polypeptides have been described as nonvesicular cytosolic ceramide transporters and renamed CERTL (GPBP) and CERT (GPBPΔ26) (5, 14). However, the conclusions of these authors were made in the absence of precise data related to the intracellular distribution of the native proteins and with complete disregard of immunochemical evidence demonstrating predominant expression of GPBP in association with basement membranes (3). More recent reports have shown that CERT-dependent ceramide transport is critical for recruitment of phospholipase A2α as well as for the recruitment and activation of protein kinase D at the trans-Golgi network, thereby ultimately regulating prostaglandin production and protein exocytosis, respectively (6, 15).
Immunohistochemical evidence suggests that GPBP is primarily extracellular, although with the potential to localize to various intracellular sites (3, 4). Protein distribution is highly informative with respect to protein function; therefore, additional studies were needed to understand the biological function of GPBP. Here we demonstrate that the translation of the mRNA for GPBP generated several polypeptides, none of which were significantly expressed in the cytosol. On the contrary, the current study provides evidence that GPBP enters into the secretory pathway and interacts with type IV collagen. Furthermore, we show that removal of the 26-residue Ser-rich region by alternative exon splicing localizes the protein to the cytosol, revealing that GPBPΔ26/CERT represents a soluble, intracellular version of GPBP. The present data suggest that alternative exon splicing and translation initiation are strategies to direct the products of COL4A3BP to different locations where they are expected to coordinate a multi-compartmental biological program. Various lines of evidence support that the latter includes phosphorylation and regulation of basement membrane collagen organization (GPBP) (1, 3, 4) and interorganelle ceramide transport that regulates vesicular protein cargo transport to the plasma membrane (GPBPΔ26/CERT) (6, 14).
MATERIALS AND METHODS
Antibodies and Recombinant Proteins—Using truncated recombinant GPBP isoforms and synthetic peptides, we have mapped the epitope of GPBP/GPBPΔ26-specific mouse monoclonal antibody (mAb) 14 (1) to the FFAT motif (supplemental Fig. S1). Mouse mAb e26 was raised against the 26 residues characteristic of GPBP (GPBPpep1) and therefore was not reactive with GPBPΔ26/CERT (see Fig. 1A). Human monoclonal F(ab)2 fragments were isolated from a recombinant F(ab)2 expression library using a synthetic peptide representing the alternatively translated region (ATR) of GPBP (see Fig. 2C) (Antibodies by Design, MorphoSys AG). Reactive F(ab)2 fragments were further characterized using Western blot and recombinant proteins expressing the predicted ATR (not shown). The most reactive F(ab)2 fragment (Ab 24) was used to characterize native GPBP polypeptides, and the least reactive F(ab)2 fragment (Ab 20) was used as negative control in these studies. The previously reported (4) immunopurified chicken polyclonal GPBP-specific antibodies (αGPBP) were biotinylated for use in flow cytometry or labeled with Alexa Fluor 647 (Invitrogen) for direct immunofluorescence. Polyclonal antibodies specific for GPBP and GPBPΔ26/CERT (αGPBPr) were produced in rabbits immunized with glutathione S-transferase-FLAG-GPBP (1) following standard procedures. Specific antibodies were affinity-purified from rabbit serum using recombinant FLAG-GPBP (see below) bound to Sepharose-CNBr (Sigma). For glyceraldehyde-3-phosphate dehydrogenase detection, we used a mouse monoclonal antibody provided by Erwin Knecht. Polyclonal antibodies specific for calregulin, p65, or cathepsin D were from Santa Cruz Biotechnology Inc., and those specific for pyruvate dehydrogenase were from Molecular Probes. Monoclonal antibodies specific for PrP (clone 3F4) or for golgin-97 were from Clontech and Molecular Probes, respectively. To detect FLAG, we used FLAG/M2 or FLAG/M2-horseradish peroxidase (Sigma) for Western blot analysis and chicken αDDDDKY (αFLAG) or goat αDDDDKY-FITC (αFLAG-FITC) for immunofluorescences (Abcam). Alexa Fluor® 488-streptavidin was from Molecular Probes, and secondary antibodies were from Promega (anti-mouse and anti-rabbit horseradish peroxidase conjugates), Jackson Immunoresearch (anti-human F(ab)2-horseradish peroxidase), and Sigma (FITC and TRITC conjugates). Recombinant FLAG-GPBP and FLAG-GPBPΔ26 were expressed in Pichia pastoris and affinity-purified as previously described (1, 3).
FIGURE 1.
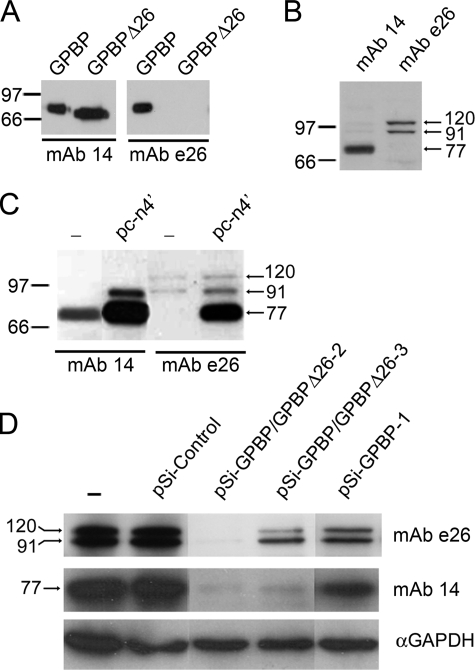
COL4A3BP encodes for polypeptides of 77-, 91-, and 120-kDa. In A, FLAG-tagged GPBP or GPBPΔ26/CERT (10–20 ng) were analyzed by Western blot with the indicated antibodies. In B, cell extracts (50 μg) were analyzed as in A. In C, extracts (10 μg) from control cells (-) or cells expressing pc-n4′were analyzed as in A. In D, extracts (50 μg) from untransfected cells (-) or from cells transfected with the indicated siRNA-expressing plasmid were analyzed as in A. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control and siRNA specificity. The reactivity of mAb e26 with native or recombinant polypeptides was fully abolished when using GPBPpep1 (20 μm) as antibody blocking peptide (not shown). In this and the following figures, numbers and bars or arrows indicate the size in kDa and the positions of the molecular mass standards or the reactive polypeptides, respectively. The results shown in this and following figures are representative of at least two independent experiments.
FIGURE 2.
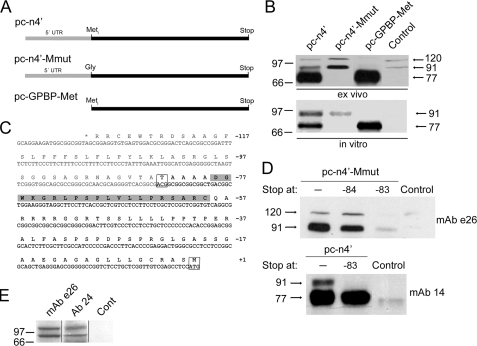
GPBP polypeptides of 91- and 120-kDa are the products of mRNA noncanonical translation initiation. In A, schematic representation of the cDNAs used to construct the indicated plasmids. In B, cell extracts (10 μg) (ex vivo) or individual transcription/translation mixtures (in vitro) expressing the indicated plasmid constructs were analyzed by Western blot using mAb e26 (ex vivo) or by fluorography (in vitro). Lysates from untransfected cells (ex vivo) or mixtures without plasmid (in vitro) were used as control. In C, indicated are the sequence of the N-terminal open reading frame of GPBP in one-letter code and the corresponding mRNA nucleotide sequence in capital letters. The gray and black letters indicate the 5′-UTR and ATR, respectively. Boxed are the codons and residues for canonical and noncanonical translation initiation. The peptide sequence targeted by Ab 24 is highlighted in gray. The negative numbers at the right denote the position of the codon or residue from canonical translation initiation site (AUG or Met, +1). In D, extracts (10 μg) from cells not expressing (Control) or expressing the indicated plasmid constructs without (-) or with a stop codon at the indicated positions were analyzed by Western blot using the indicated antibodies. In E, partially purified cell extracts (50 μg) were analyzed by Western blot using the indicated reactive species and a nonreactive F(ab)2 Ab 20 (Cont).
Plasmid Constructs—The production of pc-n4′, a pcDNA3 (Invitrogen)-derived construct expressing a cDNA that contained the 5′-untranslated region (UTR) and a coding sequence of GPBP mRNA has been reported (1). Plasmids derived from pc-n4′ included pc-GPBP-Met, a deletion mutant devoid of 5′-UTR/ATR, and pc-n4′-Mmut, a construct where the canonical AUG (Met) translation initiation was substituted with GGA (Gly). The production of pc-FLAG-GPBP, which expresses the FLAG sequence fused to the coding region of GPBP, was previously reported (1) and used to obtain pc-FLAG-GPBPΔFFAT, bearing a deletion in the FFAT motif (supplemental Fig. S1). The pc-FLAG-GPBPΔ26 expresses the FLAG sequence fused to the coding region of GPBPΔ26 and has been produced similarly to pc-FLAG-GPBP. To determine the initiation site that accounted for the ATR, we produced pc-n4′and pc-n4′-Mmut mutants by introducing stop codons at various positions in the open reading frame upstream of iMet position. The pSilence™ 2.1-U6 hygro (Ambion) was employed for transient expression of small interfering mRNAs (siRNAs) specific for GPBP or for GPBP/GPBPΔ26. The corresponding derived constructs and cDNA target sequences were: pSi-GPBP/GPBPΔ26-2, ACAGAGTATGGCTGCAGAG; pSi-GPBP/GPBPΔ26-3, GTACTTTGATGCCTGTGCT; and pSi-GPBP-1, GCCCTATAGTCGCTCTTCC. Selection of the target sequence and plasmid construction were based on the manufacturer's recommendations. The efficiency of siRNA-expressing plasmids was assessed in a cell recombinant expression system (not shown). The control plasmid in these studies (pSi-Control) was designed for targeting the mRNA of green fluorescence protein, a protein not expressed in human cells. All of the mutants were produced by standard PCR-based mutagenesis, and the fidelity of all the cDNAs cloned was confirmed by nucleotide sequencing.
Cell Culture and Transfection—HEK-293 or HeLa cells were grown with Dulbecco's modified Eagle's medium or minimal essential Eagle's medium, respectively, supplemented with 2 mm l-glutamine, 10% (v/v) fetal bovine serum and penicillin (100 units/ml)/streptomycin sulfate (0.1 mg/ml) at 37 °C in a humidified 5% CO2 environment. Unless otherwise indicated, the cells used in the studies were HEK 293 cells.
Transfections were performed for 16–24 h using ProFection mammalian transfection system-calcium phosphate (Promega) or Lipofectamine 2000 (Invitrogen), following the manufacturer's recommendations. For immunofluorescence studies, the cells were seeded on poly-l-lysine-coated coverslips in 24-well plates. When indicated HEK 293 cells were transfected with pc-n4′-Mmut and selected with G418 (Invitrogen) for 15 days. Resistant cells were further cloned by limiting dilution, and the expression of 91-kDa GPBP in a number of individual clones was determined by Western blot analysis of cell extracts (see below). Clones expressing elevated (c8, c14) or reduced (c19) levels of recombinant 91-kDa GPBP were used in functional studies.
In Vitro Transcription and Translation—We used TnT® T7-coupled reticulocyte lysate system (Promega) to perform in vitro transcription/translation of ∼1 μg of plasmid, following the manufacturer's recommendations. For assessing protein synthesis, [S35]methionine was added to the mixtures, and labeled polypeptides were identified by SDS-PAGE and fluorography. Briefly, after electrophoresis, the gels were fixed for 1 h with 45% methanol and 7.5% acetic acid. Subsequently, the gels were treated twice with dimethyl sulfoxide for 30 min and with 22.5% of 2,5-dipheniloxazol in dimethyl sulfoxide for an additional 30 min. Finally, the gels were equilibrated with water, dried, and exposed at -70 °C.
Cell Extracts and Cell Fractioning—To obtain cell extracts, growing cultures were rinsed with ice-cold phosphate bufferedsaline (PBS) and homogenized on ice bed with 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 1 mm PMSF, and 10 μg/ml leupeptin. The mixtures were cleared by centrifugation at 500 × g for 10 min, protein concentration was determined, and the mixtures were stored at -70 °C.
For subcellular fractionation, the cultures at 90% confluence were collected in PBS and subjected to centrifugation (500 × g for 10 min). Cellular pellets were dispersed in 250 mm sucrose, 10 mm PBS, pH 7.5 containing 10 μg/ml leupeptin, 1 mm PMSF and disrupted with Dounce homogenization (20 strokes) using a glass pestle. Cell homogenates were cleared progressively by sequential centrifugation to obtain the different cell fractions. The nuclei and unbroken cells were collected by centrifugation at 500 × g for 10 min. The supernatant was further cleared by centrifugation at 7,000 × g for 10 min to obtain mitochondrial/lysosome fraction. Finally, the supernatant was cleared by centrifugation at 150,000 × g for 1 h to obtain a microsomal fraction that contains fragments of cellular membranes, i.e. endoplasmic reticulum, plasma membrane, and secretory vesicles (pellet) and the cytosolic fraction (supernatant). All of the steps were performed at 0–4 °C, and the protein concentrations determined using protein assay reagent (Bio-Rad).
For some purposes, the supernatant of 500 × g was loaded on a resource-Q fast protein liquid chromatography column, and the bound material was eluted in 0–1 m NaCl gradient in 10 mm Tris-HCl, pH 8.0. The 0.55–0.6 m NaCl fractions containing the bulk of cellular GPBP were precipitated with ethanol and used as partially purified GPBP for Western blot analysis.
Ex Vivo Cross-linking, Sphingomyelinase Treatment, and FLAG Immunoprecipitation—For ex vivo cross-linking, we used HEK 293-FLAG-α3(IV) cells expressing an exportable human α3(IV)NC1 domain (BM40-FLAG-α3(IV)NC1), which was obtained essentially as previously reported (1, 16). The cells were grown up to 70–90% of confluence in either six-well plates (recombinant GPBP) or 150-mm plates (native GPBP). Cross-linking was performed 48 h after transfection or when cells reached the indicated confluence. Briefly, the cells were brought to room temperature by rinsing with PBS and incubated for 10 min with culture medium containing 1% formaldehyde. The cross-link reaction was quenched with 125 mm Gly-HCl in PBS (pH 7.4) for 10 min at room temperature. The cells were brought to 4 °C by rinsing with ice-chilled PBS and procedure continued at 4 °C. The cells were lysed with 1 or 5 ml (six-well or 150-mm plate) of extraction buffer (16 mm Tris-HCl, pH 7.5, 160 mm NaCl, 2 mm EDTA, 1.1% Triton X-100, 0.01% SDS, 10 μg/ml leupeptin, 1 mm PMSF) for 30 min, centrifuged at 500 × g for 10 min to remove cell debris, and the supernatants were extracted overnight with 50 or 250 μl (six-well or 150-mm plate) of a 50% slurry of αFLAG affinity gel using gentle rocking. The beads were collected by centrifugation and washed twice with 1 ml of extraction buffer and once with Tris-buffered saline (50 mm Tris-HCl, pH 7.5, 150 mm NaCl). The proteins were eluted twice with 25 or 125 μl (six-well or 150-mm plate) of a 100 μg/ml solution of FLAG peptide in Tris-buffered saline at room temperature. The eluted samples were boiled with electrophoresis sample buffer (twice) for 15 min to reverse cross-linking and further analyzed by SDS-PAGE and either Coomassie Blue staining or Western blot.
When indicated, HeLa cells transfected with pc-FLAG-GPBP or pc-FLAG-GPBPΔ26 were treated or not with Bacillus cereus sphingomyelinase (Sigma) as previously described (5), and cells were either fixed with methanol/acetone and analyzed by direct immunofluorescence (see below) or lysed in 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 1 mm EDTA, 50 mm NaF, 1 mm sodium orthovanadate, 10 μg/ml leupeptin, 1 mm PMSF, cleared by centrifugation (500 × g for 10 min) and used for FLAG immunoprecipitation (see above). The immunopurified materials from untreated cells were divided, and one-half was treated with 5 units/μl of λPPase (New England Biolabs) at 30 °C for 30 min following the manufacturer's recommendations. All of the samples were further analyzed by Western blot using anti-FLAG antibodies.
For some experiments, the cells were grown in 150-mm plates, transfected with 20 μg of plasmid constructs encoding FLAG-tagged proteins, and cultured for 2 additional days in fresh medium. Twenty milliliters of medium were used for FLAG immunoprecipitation essentially as indicated above.
Flow Cytometry—The cells were gently detached and dispersed in culture medium. Nonspecific antibody-binding sites on the cell surface were blocked with mouse ascites fluid containing nonrelevant mAb (blocking solution). The cells were subsequently incubated in blocking solution in the presence or absence of biotinylated αGPBP with or without blocking peptide (GPBPpep1) or with a nonrelevant synthetic peptide. The cells were incubated with Alexa Fluor® 488-streptavidin in blocking solution and further subjected to analysis in a Cytomics FC500 flow cytometer (Beckman Coulter) to measure fluorescence emission. Cell integrity was assessed measuring forward and side scattering, using untreated fresh cells as reference. All of the incubations were at room temperature for 1 h.
Direct and Indirect Immunofluorescence with Fixed Cells—The cells were transfected and fixed with methanol-acetone (1/1) chilled at -20 °C for 10 min. Subsequently, the cells were incubated with blocking solution (rabbit serum diluted 1:2 in PBS) for 30 min at room temperature, incubated with the primary antibodies (20 μg/ml in blocking solution) for 2 h at 37 °C in a humidified chamber, followed by incubation with the secondary antibody (1:200 in blocking solution) for 1 h at room temperature. The cells were stained with 4′,6′-diamino-2-phenylindole (1.25 μg/ml) in mounting fluid (DAKO) and visualized in an Axioskop-2 plus microscope (Carl Zeiss) combined with a Spot camera and software v2.2 (Diagnostic Instruments). For some experiments, the cells were transfected, fixed, incubated with αFLAG-FITC, and visualized as above indicated. Nontransfected cells were used as negative controls.
Direct Immunofluorescence of Living Cells—The cells were cultured on glass-bottomed Microwell dishes (MatTek Corp), and when they reached ∼50% confluence, the media were discarded and replaced by fresh media containing 10 μg/ml αGPBP-Alexa Fluor 647 with an excess of GPBPpep1 or equimolecular amounts of an unrelated synthetic peptide along with Rhodamine 123 (Invitrogen) for mitochondrial staining of living cells. Live cell analysis of fluorescence was performed with a Leica TCS SP2 inverted confocal microscope. The cells were maintained at 37 °C in a humidified 5% CO2 environment in all of the steps.
Mass Spectrometry—Individual protein bands were excised from Coomassie Blue-stained gel, distained, in-gel trypsin digested, and centrifuged. One microliter of the supernatant was dried and resuspended with 1 μl of matrix solution (α-cyano-4-hydroxycinnamic acid; Sigma), applied to the sample plate, dried, and introduced into the mass spectrometer. Tryptic digests peptides were analyzed by MALDI/TOF/TOF mass spectrometry (4700 Proteomics Analyzer; Applied Biosystems). The collected data were analyzed with GPS software (Applied Biosystems), and protein identification was carried out using the search engine MASCOT v 2.0 (Matrix Science).
SDS-PAGE and Western Blot Analysis—Were performed under reducing conditions following standard procedures and using chemiluminescence (Amersham Biosciences) for antibody detection.
RESULTS
COL4A3BP Encodes for Polypeptides of 77-, 91-, and 120-kDa—To identify GPBP and GPBPΔ26/CERT, we have used two different monoclonal antibodies: mAb 14, which was previously reported to recognize GPBP and GPBPΔ26/CERT (1), and mAb e26, a novel monoclonal antibody raised against the 26-residue Ser-rich region exclusive for GPBP (Fig. 1A). Using GPBP deletion mutants and synthetic peptides, we have mapped mAb 14 epitope to the FFAT motif, and thus, this antibody did not react with a GPBP mutant lacking the FFAT motif (GPBPΔFFAT) (Fig. S1).
Western blot analysis of cell extracts revealed that mAb 14 mainly recognized a single polypeptide with an apparent molecular mass of ∼77-kDa,3 whereas mAb e26 reacted with two polypeptides of ∼91- and 120-kDa (Fig. 1B). Minor and variable reactivity was also observed toward polypeptides of ∼77-, 60-, 50-, and 32-kDa with mAb e26 and against polypeptides of ∼91- and 120-kDa with mAb 14 (not shown). We found similar reactive molecular species in a number of cultured human cells including HEK 293 (Fig. 1B), human fibroblasts, HeLa, hTERT-RPE, and hTERT-BJ1 cells (not shown).
To further characterize COL4A3BP products, we compared expression of native and recombinant mRNAs (Fig. 1C). For these purposes, pc-n4′, a construct bearing the 5′-UTR and coding sequence of COL4A3BP (1, 17), was used in transient gene expression assays in cultured cells. The expression of pc-n4′ yielded three polypeptides of ∼77-, 91-, and 120-kDa that were detected by mAb e26. In contrast, only the ∼77- and 91-kDa polypeptides were significantly reactive with mAb 14. Strikingly, the most prominent mAb e26-reactive polypeptide in the recombinant lysates (77-kDa), representing the previously reported mRNA product (1), did not have a significant native counterpart.
To further determine the origin of native polypeptides, we used siRNAs specific for COL4A3BP (Fig. 1D). The expression of all three native polypeptides was reduced when expressing these siRNAs; however, siRNA specific for both GPBP and GPBPΔ26/CERT were more efficient at reducing the expression of 77-kDa polypeptide, whereas GPBP-specific siRNA reduced more effectively the expression of 91- and 120-kDa polypeptides (compare pSi-GPBP/GPBPΔ26-3 and pSi-GPBP-1). Collectively, our data suggested that major cellular products of COL4A3BP included GPBPΔ26/CERT (77-kDa) and the previously unrecognized GPBP isoforms of 91- and 120-kDa, the latter likely bearing a modified FFAT motif that prevented consistent mAb 14 binding. The reduction in the cellular levels of 77-kDa polypeptide when using GPBP-specific siRNAs requires further investigation because this polypeptide displayed no significant reactivity with mAb e26 (Fig. 1B).
Major Cellular GPBP Isoforms Result from Noncanonical mRNA Translation Initiation—To further define the origin of cellular GPBP isoforms, we produced (pc-n4′)-derived constructs expressing mRNA mutants consisting of 5′-UTR deletion or iMet to Gly substitution (Fig. 2A), and these were used in protein expression assays (Fig. 2B). In cells, the construct representing 5′-UTR-deleted mRNA (pc-GPBP-Met) produced only the 77-kDa polypeptide, and the constructs representing the iMet to Gly substitution (pc-n4′-Mmut) expressed only the 91- and 120-kDa polypeptides (Fig. 2B, ex vivo). However, in a cell-free translation system, pc-GPBP-Met also expressed 77-kDa GPBP polypeptide, but pc-n4′Mmut yielded only the 91-kDa polypeptide, and no significant expression of 120-kDa polypeptide was observed (Fig. 2B, in vitro). These data indicated that GPBP mRNA contained a noncanonical translation initiation site(s) in the 5′-UTR that accounted for polypeptides of 91- and 120-kDa, whereas the 77-kDa polypeptide was the product of canonical translation initiation. Moreover, our data also suggested that the 91-kDa was the primary product of noncanonical translation initiation, and the 120-kDa polypeptide represented a post-translational derived product that could not be expressed in a cell-free system devoid of cellular membranes.
To characterize further noncanonical translation initiation, the previously recognized (1) open reading frame present in the 5′-UTR of the GPBP mRNA (Fig. 2C) was interrupted by introducing a stop codon at individual positions in pc-n4′Mmut and cellular protein expression assessed by Western blot (Fig. 2D). The construct bearing a stop codon at -83 (originally ACG, threonine) did not express the 91- and 120-kDa polypeptides, but the construct with the stop codon at -84 (originally GCG, alanine) expressed the two polypeptides mapping the alternative translation start site to codon -83 (boxed Thr in Fig. 2C). The same conclusion was obtained when we assayed the -83 stop mutant of pc-n4′ (Fig. 2D).
To confirm that noncanonical translation initiation also accounted for endogenous GPBP polypeptides of 91- and 120-kDa, a human F(ab)2 fragment (Ab 24) specifically reacting with a synthetic peptide representing the predicted ATR (shaded sequence in Fig. 2C) was used for Western blot analysis of partially purified GPBP polypeptides (Fig. 2E). As expected, Ab 24 specifically reacted with two polypeptides of 91- and 120-kDa, which were also recognized by mAb e26, suggesting that native GPBP polypeptides contained the ATR characteristic of noncanonical translation products.
The 91- and 120-kDa GPBP Isoforms Are Insoluble Membrane-bound Polypeptides—GPBP isoform of 91-kDa was predicted to be nonclassical secreted proteins when analyzed with SecretomeP 2.0 Server (18) and to localize in mitochondria (60.9%), nucleus (26.1%), cytoskeleton (8.7%), and vesicles of secretory system (4.3%) when analyzed with PSORT II Prediction. Thus, these theoretical considerations suggested that GPBP isoforms resulting from noncanonical translation initiation were noncytosolic polypeptides that entered into cellular organelles including the secretory pathway.
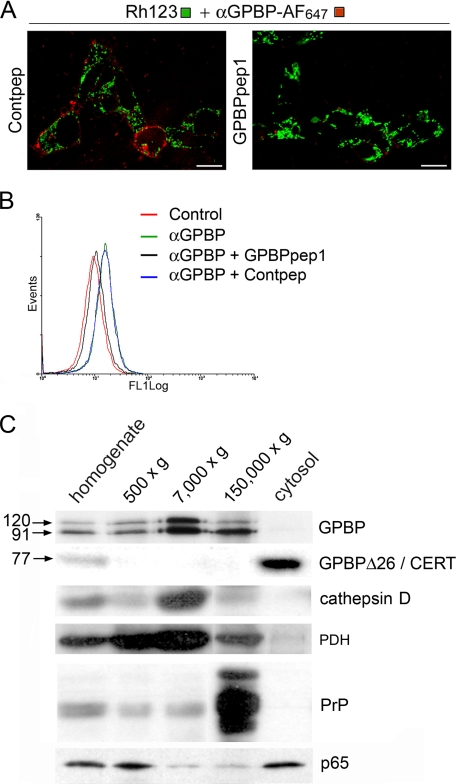
To assess these predictions, intact living cells were incubated with αGPBP and analyzed by direct immunofluorescence and flow cytometry for antibody binding detection (Fig. 3, A and B). Interestingly, αGPBP bound to living cells in a specific manner because binding of the antibodies was efficiently abolished by a synthetic peptide representing GPBP (GPBPpep1) but not by an unrelated polypeptide (Contpep). These data suggested that cellular GPBP isoforms were present in the external surface of the plasma membrane.
FIGURE 3.
The 91- and 120-kDa GPBP isoforms are insoluble membrane-bound polypeptides. In A, intact cells were incubated with αGPBP-Alexa Fluor 647 antibodies (αGPBP-AF647) in the presence of GPBPpep1 or equimolecular amount of a nonrelevant peptide (Contpep) and Rhodamine 123 for mitochondrial staining of living cells and analyzed by confocal microscopy. Scale bar, 21 μm. In B, the cells were detached and incubated with blocking solution in the absence (control) or presence of biotinylated αGPBP antibodies (αGPBP). The cell surface-bound antibody was detected with streptavidin-FITC and flow cytometry. As a control, parallel cultures were incubated with the same antibodies in the presence of GPBPpep1 (αGPBP + GPBPpep1) or equimolecular amount of a nonrelevant peptide (αGPBP + Contpep) and similarly analyzed. In C, similar amounts (10 μg) of the indicated cellular fractions were analyzed by Western blot using antibodies for the indicated proteins. We used as cellular compartment markers: pyruvate dehydrogenase (PDH) for mitochondria; cathepsin D for lysosome; prion protein (PrP) for microsome; and nuclear factor κB (p65) for nucleus and cytosol. For GPBP and GPBPΔ26/CERT detection, we used mAb e26 and mAb 14, respectively. Because we did not detect expression of 77-kDa GPBP in the cytosol, mAb 14 reactivity in this compartment can be attributed to GPBPΔ26/CERT.
To further characterize the intracellular distribution of GPBP, the cells were disrupted and subjected to subcellular fractionation and Western blot analysis (Fig. 3C). Consistent with predictions, GPBP isoforms of 91- and 120-kDa were not detected as soluble materials, but rather they were found mainly associated with mitochondrial-lysosomal (7,000 × g) and microsomal fractions (150,000 × g). It remained to be determined whether the presence of GPBP in the nuclear fraction (500 × g) indeed reflected nuclear expression of these proteins or rather unbroken cells and/or mitochondria contaminating this fraction. In contrast, a polypeptide of ∼77 kDa, which reacted with mAb 14 and showed no significant reactivity with mAb e26, was exclusively detected as soluble after sample centrifugation at 150,000 × g for 1 h (cytosol).
These data suggested that native GPBP polypeptides of 91- and 120-kDa were expressed insoluble associated with cellular membranes, whereas native GPBPΔ26/CERT polypeptide of 77-kDa was expressed soluble in the cytoplasm.
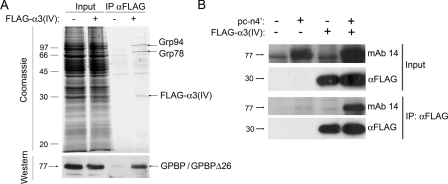
The 77-kDa GPBP Is a Soluble Extracellular Protein That Interacts with Type IV Collagen—Previous reports suggested that 77-kDa GPBP interacts with type IV collagen (1, 3, 4). This was further assessed by ex vivo cross-linking and FLAG immunoprecipitation of cells expressing BM40-FLAG-α3(IV)NC1, a recombinant exportable form of the human α3(IV)NC1 (16), followed by SDS-PAGE analysis of immunoprecipitates (Fig. 4A). FLAG-specific antibodies efficiently precipitated FLAG-α3(IV)NC1 and a 77-kDa polypeptide representing either GPBP or GPBPΔ26/CERT4 (Western) along with Grp78 and Grp94 (Coomassie), two ER-resident chaperones implicated in protein folding and ER homeostasis maintenance (19, 20). To further determine that GPBP indeed interacted with FLAG-α3(IV) in the ER, cells expressing or not expressing BM40-FLAG-α3(IV)NC1 were transfected with pc-n4′ and similarly analyzed (Fig. 4B). FLAG antibodies efficiently precipitated 77-kDa GPBP from cells expressing FLAG-α3(IV)NC1 but not from control cells, suggesting that the 77-kDa GPBP isoform enters into the secretory pathway and interacts with FLAG-α3(IV)NC1.
FIGURE 4.
The 77-kDa GPBP isoform interacts with type IV collagen in cultured cells. In A, HEK 293 or HEK 293-FLAG-α3(IV) cells were cross-linked, lysed, and αFLAG extracted. Fifty micrograms of cell lysate (Input) or the corresponding FLAG-immunoprecipitated materials (IPαFLAG) were reversed cross-linked and analyzed by Coomassie Blue staining or Western blot with αGPBPr. The major specific polypeptides in FLAG-immunoprecipitates (arrows) were excised and identified by MALDI/TOF/TOF mass spectrometry. In B, HEK 293 (-) or HEK 293-FLAG-α3(IV) (+) cells were transfected with pcDNA3 (-) or with pc-n4′ (+), cross-linked, processed, and analyzed as in A by Western blot using the indicated antibodies.
Primary structure analysis predicted a cytoplasmic localization for 77-kDa GPBP polypeptide.5 However, in vitro (1, 3), ex vivo (Fig. 4), and in vivo (4) studies suggested that the 77-kDa GPBP isoform binds and phosphorylates type IV collagen. Furthermore, although recombinant expression studies revealed that the 77-kDa GPBP polypeptide was the most prominent polypeptide, no significant levels of the native counterpart were detected within the cells (Fig. 1). Collectively, these observations suggested that canonical GPBP was a cytosolic polypeptide subjected to a nonclassical secretion.
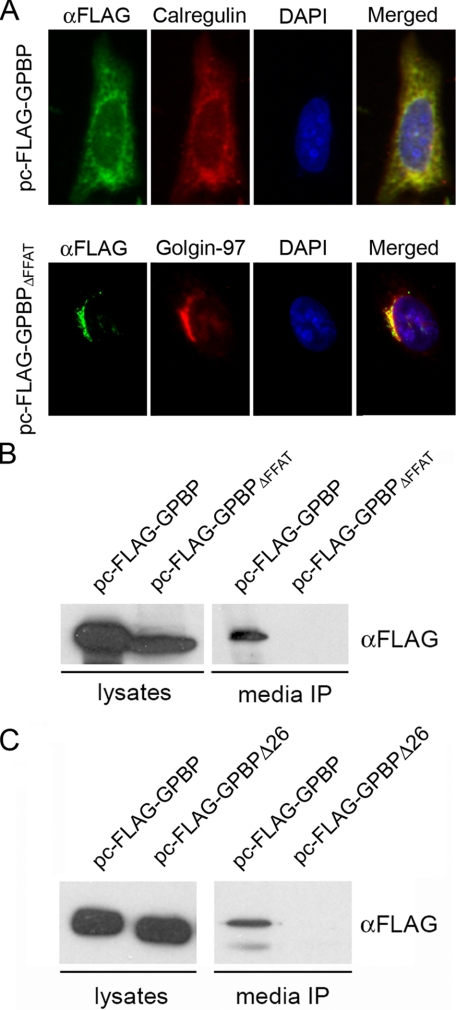
To explore whether GPBP was secreted, we first expressed FLAG-tagged GPBP in HeLa cells and used FLAG-specific antibodies to analyze intracellular recombinant protein distribution (Fig. 5A). FLAG-GPBP co-localized extensively with calregulin, an ER-resident protein, suggesting that, as described for GPBPΔ26/CERT (21, 22), FLAG-GPBP bound to the ER through FFAT-VAP interaction. Consequently, we expressed and similarly analyzed FLAG-GPBPΔFFAT, a FLAG-GPBP variant devoid of FFAT motif. Deletion of FFAT motif prevented distribution of GPBP to the ER, because the protein was found extensively co-localizing with golgin-97, a Golgi apparatus resident protein (Fig. 5A). Identical conclusions were obtained when the studies were conducted in HEK 293 cells (not shown). Our data were consistent with the notion that recombinant GPBP was a cytosolic protein bound to VAP through the FFAT motif for its exportation, and only when FFAT interaction was impaired did the protein have the potential to associate with Golgi apparatus. This was explored by expressing FLAG-GPBP or FLAG-GPBPΔFFAT in cultured cells and the subsequent analysis of culture media by immunoprecipitation and Western blot analysis (Fig. 5B). Interestingly, FLAG-specific antibodies efficiently immunoprecipitated recombinant protein from the media of cultures expressing FLAG-GPBP but not from the media of cells expressing FLAG-GPBPΔFFAT, revealing that FFAT-mediated binding to the ER is essential for 77-kDa GPBP secretion.
FIGURE 5.
Export of 77-kDa GPBP to the extracellular compartment. In A, HeLa cells were transfected with the indicated plasmid constructs, and the indicated proteins were visualized by standard indirect immunofluorescence. DNA was stained with 4′,6′-diamino-2-phenylindole (DAPI) for nuclear visualization. Original magnification, ×400. In B and C, extracts (10 μg) from cells expressing the indicated plasmid constructs (lysates) or FLAG immunoprecipitates from the corresponding culture media (media IP) were analyzed by Western blot using the indicated antibodies.
GPBPΔ26/CERT also binds to the ER in a FFAT-dependent manner (21, 22); however, we found GPBPΔ26/CERT in the cytosol (Fig. 3C) and 77-kDa GPBP in the extracellular compartment (Fig. 5B), supporting the idea that the Ser-rich 26-residue region exclusive to GPBP is also critical for GPBP secretion. This was similarly explored in cultures expressing FLAG-tagged 77-kDa GPBP or GPBPΔ26/CERT (Fig. 5C). As expected, the presence of the 26-residue Ser rich region was critical for protein secretion given that FLAG-GPBPΔ26 was not significantly expressed in the culture medium.
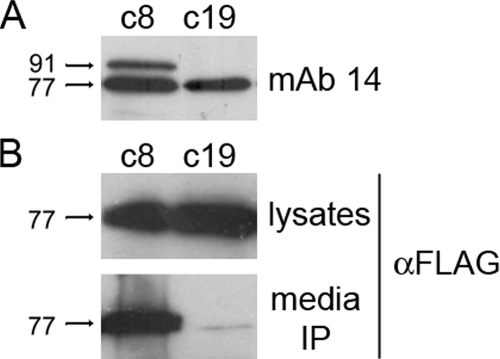
The 91-kDa GPBP Regulates the Levels of 77-kDa GPBP in the Extracellular Compartment—The evidence supports that both the 77- and 91-kDa GPBP isoforms enter into the secretory pathway, but whereas the 91-kDa remains associated to membranes, the 77-kDa GPBP is soluble in the extracellular compartment. We have explored whether 91-kDa GPBP regulated the extracellular levels of 77-kDa GPBP. This was accomplished by recombinant expression of FLAG-GPBP in individual cell lines expressing recombinant 91-kDa GPBP to different levels (Fig. 6A) followed by FLAG immunoprecipitation of the corresponding cultured medium and analysis of immunoprecipitates by Western blot (Fig. 6B). Interestingly, increased recombinant 91-kDa GPBP expression associated with increased levels of FLAG-GPBP in the culture medium, suggesting that 91-kDa GPBP induced the secretion of 77-kDa GPBP to the extracellular compartment.
FIGURE 6.
The 91-kDa GPBP regulates 77-kDa GPBP secretion in cultured cells. In A, extracts (10 μg) from two independent clones expressing (c8) or not expressing (c19) recombinant 91-kDa GPBP were analyzed by Western blot with mAb 14 antibodies, which react poorly with native 91-kDa counterpart (Fig. 1B). In B, the same clones were transfected with pc-FLAG-GPBP, and cell extracts (lysates) or FLAG immunoprecipitates from the corresponding culture media (media IP) were analyzed by Western blot using the indicated antibodies. Similar conclusions were obtained when assaying c14, an independent HEK 293 clone expressing levels of recombinant 91-kDa GPBP similar to c8 (not shown).
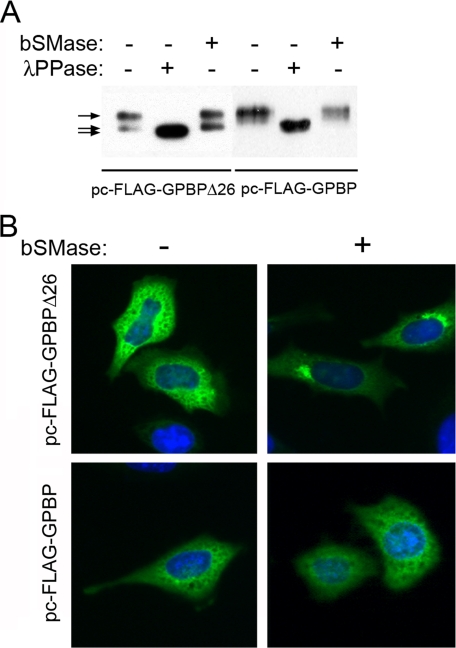
The 77-kDa GPBP Is Not Sensitive to Cell Treatment with Sphingomyelinase—Recombinant expression studies also showed that 77-kDa GPBP was a cytosolic polypeptide associated with ER that underwent translocation to the Golgi apparatus when FFAT motif was mutated (Fig. 5A). Consequently, we asked whether 77-kDa GPBP underwent dephosphorylation and translocation to the Golgi apparatus in response to sphingomyelinase cell treatment as previously reported for GPBPΔ26/CERT (5). For these studies, cells expressing FLAG-tagged GPBP or GPBPΔ26/CERT were treated with B. cereus sphingomyelinase, and intracellular proteins of interest were analyzed by FLAG immunoprecipitation and Western blot (Fig. 7A). As previously noted (1, 5), both recombinant proteins were phosphorylated, and incubation with a dual specificity phosphatase (λPPase) reduced their molecular mass to a similar extent (top and bottom arrows). However, sphingomyelinase cell treatment had different consequences for each recombinant protein; whereas the relative amount of FLAG-GPBPΔ26/CERT, which displayed lower molecular mass (Fig. 7A, middle arrow) increased from ∼25% to ∼50%, no significant variation in the electrophoretic pattern of FLAG-GPBP was observed. This suggested that the reduction in the cellular levels of sphingomyelin caused by sphingomyelinase treatment induced partial dephosphorylation of FLAG-GPBPΔ26/CERT but did not affect significantly the phosphorylation state of FLAG-GPBP. As expected (5), immunofluorescence analysis of the cells revealed that sphingomyelinase treatment promoted translocation of FLAG-GPBPΔ26/CERT to the Golgi apparatus but did not alter significantly the intracellular distribution of FLAG-GPBP (Fig. 7B).
FIGURE 7.
GPBPΔ26/CERT but not GPBP is sensitive to sphingomyelinase cell treatment. In A, HeLa cells were transfected with the indicated plasmid constructs and treated (+) or not (-) with spingomyelinase, lysed, FLAG immunoprecipitated, and analyzed by Western blot with αFLAG antibodies (bSMase). Immunoprecipitates from untreated cells were incubated (+) or not (-) with phosphatase and similarly analyzed (λPPase). We have used a 8–12% gradient gel and extensive electrophoresis to separate phosphorylated and dephosphorylated versions of FLAG-tagged GPBPΔ26/CERT and estimated their relative abundance by Western blot and densitometry. In B, the same cells as in A were fixed by methanol/acetone, double-labeled with anti-FLAG-FITC antibody (green) and 4′,6′-diamino-2-phenylindole (blue), and analyzed by direct immunofluorescence. Original magnification, ×400.
DISCUSSION
Here we have obtained compelling evidence that the mRNA of GPBP undergoes canonical (AUG) and noncanonical (ACG) translation initiation to generate two primary polypeptides of 77 and 91 kDa, respectively. The results from the present study also support that both products enter the secretory pathway. However, whereas the 77-kDa polypeptide reaches the extracellular compartment and exists in a soluble immunoprecipitable form, the 91-kDa polypeptide and its derived 120-kDa polypeptide remain mainly insoluble, associated with cellular membranes. The use of translation initiation at ACG and non-canonical translation initiation to direct proteins to alternative cell compartments have been described for other human genes (23, 24). Based on previous evidence (21, 22), it is expected that FFAT-mediated GPBP binding to the ER (Fig. 5) occurs through VAP and therefore that FFAT-VAP interaction mediates molecular mechanisms underlying GPBP translocation into the ER. Furthermore, we also show that the previously reported alternatively spliced GPBPΔ26/CERT is a GPBP variant that remains mainly soluble in the cytoplasm. Thus, our data support the notion that mRNA alternative translation initiation and exon splicing are strategies to direct GPBP to multiple locations including the cytosol, secretory pathway, plasma membrane, and extracellular compartment. Moreover, previous observations have localized GPBP to the nucleus in human spermatogonium (1) and in the mitochondria and lysosome of rat liver,6 suggesting that the distribution of GPBP is virtually ubiquitous, and therefore its biological program is expected to be exerted in several compartments.
A human GPBP cDNA from pulmonary artery endothelial cell has been reported (GenBank™ accession number AK096854). Interestingly, AK096854 bears an alternative canonical translation initiation site (iMet) that extends the open reading frame of the 91-kDa polypeptide upstream by 45 residues. We have not found evidence for AK096854 mRNA expression in HEK 293 cells, nor in a number of other human tissues including liver, kidney, brain, muscle, pancreas, keratinocytes, lymphocytes, and HeLa cells (not shown). Nevertheless, the existence of GPBP isoforms produced by canonical mRNA translation initiation (i.e. AK096854) with a molecular mass similar to that of the noncanonical translation initiation products reported here cannot be excluded.
Primary structure analysis predicts that noncanonically translated GPBP products enter into the secretory pathway. Several observations support these predictions, namely: 1) non-canonical GPBP isoforms are molecular species associated with cellular membranes (Fig. 3); 2) noncanonical GPBP isoforms are the predominant GPBP species in the cell (Fig. 1), and GPBP-specific antibodies bound to the external surface of intact living cells (Fig. 3); 3) the 120-kDa GPBP polypeptide is not expressed from the mRNA when translation occurs in a cell-free system devoid of cellular membranes (Fig. 2); and 4) the 91-kDa GPBP isoform regulates the levels of the 77-kDa GPBP at the extracellular compartment (Fig. 6). Taken together, these observations support the notion that the 91-kDa polypeptide is the primary product of noncanonical translation initiation. This isoform enters into the secretory pathway where it undergoes covalent modification to yield the 120-kDa polypeptide and remains bound to membranes reaching the external surface of the plasma membrane. The mechanism by which 91-kDa GPBP regulates the extracellular levels of 77-kDa GPBP remains unknown.
We have observed that when expression is abnormally elevated (i.e. transient gene expression), GPBP polypeptides accumulate in the cytosol (supplemental Fig. S2), revealing that GPBP transportation into the ER is a saturable process. Interestingly, under these expression conditions, mAb e26 displayed more reactivity for the cytosolic 77-kDa polypeptide than for this isoform when residing in the extracellular compartment (supplemental Figs. S2 and S3). Moreover, mAb 14 reacted comparatively more with recombinant than with native 91-kDa GPBP and did not react significantly with native or recombinant 120-kDa product (Fig. 1). All of these observations suggest that the 26-residue Ser-rich region (mAb e26) and the FFAT motif (mAb 14) are subjected to covalent modifications in the secretory pathway. These data also imply that under specific regulatory (physiological or pathological) circumstances, GPBP can be expressed as soluble polypeptides in the cytosol. Finally, it remains to be determined whether 91-kDa GPBPΔ26/CERT is expressed endogenously and whether GPBPΔ26/CERT can be transported into the ER without undergoing secretion.
The expression levels of cytosolic 77-kDa polypeptide representing GPBPΔ26/CERT were significantly reduced in cells expressing GPBP-specific siRNA (Fig. 1D). This suggests that either siRNA is also targeting the pre-mRNA or that the mRNA of GPBP is to some extent a precursor of GPBPΔ26 mRNA. We have found that cells expressing recombinant GPBP mRNA also expressed limited amounts of recombinant GPBPΔ26/CERT mRNA.5 This suggests that mature GPBP mRNA is subjected to a nonclassical processing, similarly to that reported for XBP1 in response to ER stress signals (25). Alternatively, GPBP species bearing a covalently modified 26-residue Ser-rich region that co-migrate with GPBPΔ26/CERT could also account for this observation.
Several lines of evidence support the idea that GPBP regulates protein folding in the ER and supramolecular organization in the extracellular compartment rather than interorganelle ceramide traffic in the cytosol: 1) the 77-kDa GPBP is a non-conventional Ser/Thr kinase that binds and phosphorylates the α3(IV)NC1 domain at sites (1) that are also phosphorylated in vivo (26); 2) the 77-kDa GPBP is mainly found in the extracellular compartment both soluble (Fig. 5) or associated with glomerular basement membrane collagen (4) and is not expressed at significant levels in the cytosol of cultured cells (Figs. 1 and 3); 3) cellular GPBP isoforms localize at the external surface of the plasma membrane (Fig. 3); 4) the 91-kDa GPBP isoform is associated with cellular membranes (Fig. 3) and regulates the extracellular levels of the 77-kDa GPBP isoform (Fig. 6); 5) the α3(IV)NC1 domain undergoes unique structural diversification, and at least two distinct conformational isoforms (conformers) assemble in basement membranes (27); 6) an increased expression of the 77-kDa GPBP perturbs the quaternary structure of type IV collagen, suggesting that the elevated GPBP levels interfere with the conformational diversification program (tertiary structure) of the α3(IV)NC1 domain (4); 7) the FFAT motif is a structural requirement for 77-kDa GPBP secretion (Fig. 5), and VAP is critical for maintaining the homeostasis for adequate protein folding in the ER (10); 8) Grp78 and Grp94, chaperones that reside in the ER and regulate cellular response to protein misfolding (18, 19), are associated with FLAG-α3(IV) and 77-kDa GPBP (Fig. 4); 9) increased COL4A3BP expression has been found to mediate resistance of cancer cells to chemotherapeutic agents that induce protein misfolding and ER stress-mediated cell death (28); 10) treatment of cells with sphingomyelinase does not induce dephosphorylation nor does it alter intracellular distribution of 77-kDa GPBP (Fig. 7); 11) protein kinase D phosphorylates GPBP but not to the same extent as GPBPΔ26/CERT (6); 12) knock-down and rescue experiments reveal that GPBP and GPBPΔ26/CERT exert different biological functions during embryogenesis in Zebra fish (29); and 13) GPBP interacts with proteins RTN3 and RTN4, which are anchored from the luminal/extracellular side to the membranes in the secretory pathway (30). GPBP lacking the 26-residue Ser-rich region also binds to VAP (21, 22); however, ceramide uptake follows binding to VAP, and subsequently the protein departs to the Golgi apparatus where ceramide is released and protein exocytosis is induced (6, 14). Therefore, phosphate transfer and ceramide trafficking may be molecular strategies through which COL4A3BP regulates protein secretion (i.e. type IV collagen). Consistent with this, it has been shown that VAP is also critical for regulating protein cargo transport to the plasma membrane (11).
Various lines of evidence support that COL4A3BP is an attractive target for strategies to diagnose and treat antibody-mediated disorders (3, 4), inflammation (15), ER stress-mediated diseases (10), and drug-resistant cancer (28). However, observations supporting these conclusions may now need to be reinterpreted because many have been obtained using tools (i.e. siRNA or antibodies) that failed to discriminate between different gene products (i.e. GPBP and GPBPΔ26/CERT), which are expressed at distinct cell compartments and are differentially regulated in response to stimuli (3). Therefore, the present study makes an important contribution to this understanding by clarifying the mechanisms by which various isoforms of GPBP are generated within the cells.
Supplementary Material
Acknowledgments
We thank Natalia Salvador and Erwin Knecht for assistance in performing in vitro translation assays. The technical assistance of Andreu Jordán and Zahara Garzón and of Alberto Hernandez-Cano from the Servicio de Confocal (Centro de Investigación Príncipe Felipe, Fondo Europeo de Desarrollo Regional) is greatly appreciated. Proteomic analysis has been performed in the Unidad de Proteómica at Centro de Investigación Príncipe Felipe, which is member of ProteoRed. We also want thank Deborah Burks for the critical reading of the manuscript.
This work was supported by Grants SAF97/0065, SAF2000/0047, SAF2001/0453, SAF2003-09772-C03-01, and SAF2006-12520-C02-01 from Ministerio de Educación y Ciencia, Grant 98/102-00 from Fundación “La Caixa,” and Grants GV04B-285 and BM-001/2002 from Generalitat Valenciana (Spain) (to J. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: GPBP, Goodpasture antigen-binding protein; GPBPΔ26, short alternatively spliced variant of GPBP; ATR, alternative translated region; CERT and CERTL, short and large isoforms of the ceramide transfer protein; ER, endoplasmic reticulum; FFAT, two phenylalanines in an acidic tract; mAb, monoclonal antibody; NC1, noncollagenous-1 domain; PBS, phosphate buffered-saline; PMSF, phenylmethylsulfonyl fluoride; START, steroidogenic acute regulatory related lipid transfer; UTR, untranslated region; VAP, vesicle associated membrane protein-associated protein; Ab, antibody; FITC, fluorescein isothiocyanate; TRITC, tetramethylrhodamine isothiocyanate; siRNA, small interfering RNA; MALDI/TOF/TOF, matrix-assisted laser desorption ionization time-of-flight time-of-flight.
The 77-kDa polypeptide can be resolved as a doublet representing phosphorylated (higher) and dephosphorylated (lower) versions of GPBPΔ26/CERT (5).
Secretion of 77-kDa GPBP associated with the loss of reactivity with mAb e26 (supplemental Fig. S3), excluding the use of this antibody to estimate the levels of native 77-kDa GPBP in the secretory pathway.
F. Revert, I. Ventura, P. Martínez-Martínez, F. Granero-Moltó, F. Revert-Ros, J. Macías, and J. Saus, unpublished observations.
P. Martínez-Martínez, N. Salvador, E. Knecht, and J. Saus, unpublished observations.
References
- 1.Raya, A., Revert, F., Navarro, S., and Saus, J. (1999) J. Biol. Chem. 274 12642-12649 [DOI] [PubMed] [Google Scholar]
- 2.Hudson, B. G., Tryggvason, K., Sundaramoorthy, M., and Neilson, E. G. (2003) N. Engl. J. Med. 348 2543-2556 [DOI] [PubMed] [Google Scholar]
- 3.Raya, A., Revert-Ros, F., Martínez-Martínez, P., Navarro, S., Roselló, E., Vieites, B., Granero, F., Forteza, J., and Saus, J. (2000) J. Biol. Chem. 275 40392-40399 [DOI] [PubMed] [Google Scholar]
- 4.Revert, F., Merino, R., Monteagudo, C., Macías, J., Peydró, A., Alcácer, J., Muniesa, P., Marquina, R., Blanco, M., Iglesias, M., Revert-Ros, F., Merino, J., and Saus, J. (2007) Am. J. Pathol. 171 1419-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumagai, K., Kawano, M., Shinkai-Ouchi, F., Nishijima, M., and Hanada, K. (2007) J. Biol. Chem. 282 17758-17766 [DOI] [PubMed] [Google Scholar]
- 6.Fugmann, T., Hausser, A., Schöffler, P., Schmid, S., Pfizenmaier, K., and Olayioye, M. A. (2007) J. Cell Biol. 178 15-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemmon, M. A., and Ferguson, K. M. (2000) Biochem. J. 350 1-18 [PMC free article] [PubMed] [Google Scholar]
- 8.Dowler, S., Currie, R. A., Campbell, D. G., Deak, M., Kular, G., Downes, C. P., and Alessi, D. R. (2000) Biochem. J. 351 19-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewen, C. J. R., Roy, A., and Levine, T. P. (2003) EMBO J. 22 2025-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanekura, K., Nishimoto, I., Aiso, S., and Matsuoka, M. (2006) J. Biol. Chem. 281 30223-30233 [DOI] [PubMed] [Google Scholar]
- 11.Wyles, J. P., McMaster, C. R., and Ridgway, N. D. (2002) J. Biol. Chem. 277 29908-29918 [DOI] [PubMed] [Google Scholar]
- 12.Soccio, R. E., and Breslow, J. L. (2003) J. Biol. Chem. 278 22183-22186 [DOI] [PubMed] [Google Scholar]
- 13.Alpy, F., and Tomasetto, C. (2005) J. Cell Sci. 118 2791-2801 [DOI] [PubMed] [Google Scholar]
- 14.Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., and Nishijima, M. (2003) Nature 426 803-809 [DOI] [PubMed] [Google Scholar]
- 15.Lamour, N. F., Stahelin, R. V., Wijesinghe, D. S., Maceyka, M., Wang, E., Allegood, J. C., Merrill, A. H. Jr., Cho, W., and Chalfant, C. E. (2007) J. Lipid Res. 48 1293-1304 [DOI] [PubMed] [Google Scholar]
- 16.Netzer, K. O., Leinonen, A., Boutaud, A., Borza, D. B., Todd, P., Gunwar, S., Langeveld, J. P., and Hudson, B. G. (1999) J. Biol. Chem. 274 11267-11274 [DOI] [PubMed] [Google Scholar]
- 17.Granero, F., Revert, F., Revert-Ros, F., Laínez, S., Martínez-Martínez, P., and Saus, J. (2005) FEBS J. 272 5291-5305 [DOI] [PubMed] [Google Scholar]
- 18.Bendtsen, D. J., Jensen, J. L., Blom, N., von Heijne, G., and Brunak, S. (2004) Protein Eng. Des. Sel. 17 349-356 [DOI] [PubMed] [Google Scholar]
- 19.Yang, Y., and Li, Z. (2005) Mol. Cell 20 173-18216246721 [Google Scholar]
- 20.Ni, M., and Lee, A. S. (2007) FEBS Lett. 581 3641-3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry, R. J., and Ridgway, N. D. (2006) Mol. Biol. Cell 17 2604-2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawano, M., Kumagai, K., Nishijima, M., and Hanada, K. (2006) J. Biol. Chem. 281 30279-30288 [DOI] [PubMed] [Google Scholar]
- 23.Peabody, D. S. (1989) J. Biol. Chem. 264 5031-5035 [PubMed] [Google Scholar]
- 24.Touriol, C., Bornes, S., Bonnal, S., Audigier, S., Prats, H., Prats, A. C., and Vagner, S. (2003) Biol. Cell. 95 169-178 [DOI] [PubMed] [Google Scholar]
- 25.Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001) Cell 107 881-891 [DOI] [PubMed] [Google Scholar]
- 26.Revert, F., Penadés, J. R., Plana, M., Bernal, D., Johansson, C., Itarte, E., Cervera, J., Wieslander, J., Quinones, S., and Saus, J. (1995) J. Biol. Chem. 270 13254-13261 [DOI] [PubMed] [Google Scholar]
- 27.Calvete, J. J., Revert, F., Blanco, M., Cervera, J., Tárrega, C., Sanz, L., Revert-Ros, F., Granero, F., Pérez-Payá, E., Hudson, B. G., and Saus, J. (2006) Proteomics 6 S237-S244 [DOI] [PubMed] [Google Scholar]
- 28.Swanton, C., Marani, M., Pardo, O., Warne, P. H., Kelly, G., Sahai, E., Elustondo, F., Chang, J., Temple, J., Ahmed, A. A., Brenton, J. D., Downward, J., and Nicke, B. (2007) Cancer Cell 11 498-512 [DOI] [PubMed] [Google Scholar]
- 29.Granero-Moltó, F., Sarmah, S., O'Rear, L., Spagnoli, A., Abrahamson, D., Saus, J., Hudson, B. G., and Knapik, E. W. (2008) J. Biol. Chem. 283 20495-20504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rual, J. F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., Berriz, G. F., Gibbons, F. D., Dreze, M., Ayivi-Guedehoussou, N., Klitgord, N., Simon, C., Boxem, M., Milstein, S., Rosenberg, J., Goldberg, D. S., Zhang, L. V., Wong, S. L., Franklin, G., Li, S., Albala, J. S., Lim, J., Fraughton, C., Llamosas, E., Cevik, S., Bex, C., Lamesch, P., Sikorski, R. S., Vandenhaute, J., Zoghbi, H. Y., Smolyar, A., Bosak, S., Sequerra, R., Doucette-Stamm, L., Cusick, M. E., Hill, D. E., Roth, F. P., and Vidal, M. (2005) Nature 437 1173-1178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.