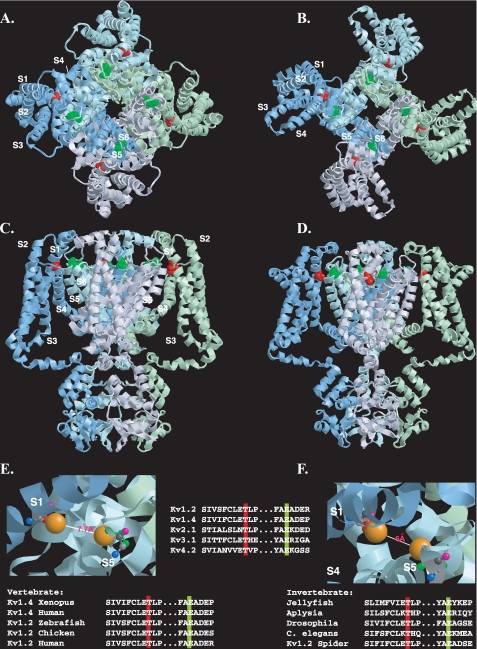
FIGURE 9.
Disposition of the conserved threonine residue within the resting and open state structural models of Kv1.2. A-D, view of the Kv1.2 tetramer in resting (A and C) and open (B and D) state models (7) shown from the extracellular side of the membrane (A and B) or from the side of the membrane (C and D). For clarity, each subunit monomer has been assigned a separate color and displayed schematically. Transmembrane-spanning regions are labeled S1-S6. The conserved threonine (Thr-184) and glutamate (Glu-350) residues (corresponding to Thr-330 and Glu-502 in Kv1.4), are colored red and green, respectively. E and F, close-up view of the putative interlocking region, showing Thr-184 and Glu-350 in ball-and-stick representation for resting (E) and open (F) state models, with side chains (Thr-184, red; Glu-350, green) extended but no α-chain rotation. Oxygen atoms involved in potential hydrogen bonding are shown in orange. Backbone nitrogen, carbonyl oxygen, andα andβ carbons are denoted in blue, pink, light gray, and dark gray, respectively. Top inset, disposition of transmembrane regions colored as follows: S1, yellow; S2, orange; S3, purple; S4, pink; S5, green; S6, blue. Lower insets, evolutionary conservation of the extracellular threonine and glutamate residues colored according to the scheme described in A-D. Modeling was done using RasTop, a Windows interface to RasMol 2.62 (originally written by Roger Sayle). Structural coordinates for the resting and open state were obtained from the supplementary data given in Ref. 7 and correspond to a refinement of the original Kv1.2 crystal structure (6, 21) using the Rosetta-Membrane method and molecular dynamics simulations.