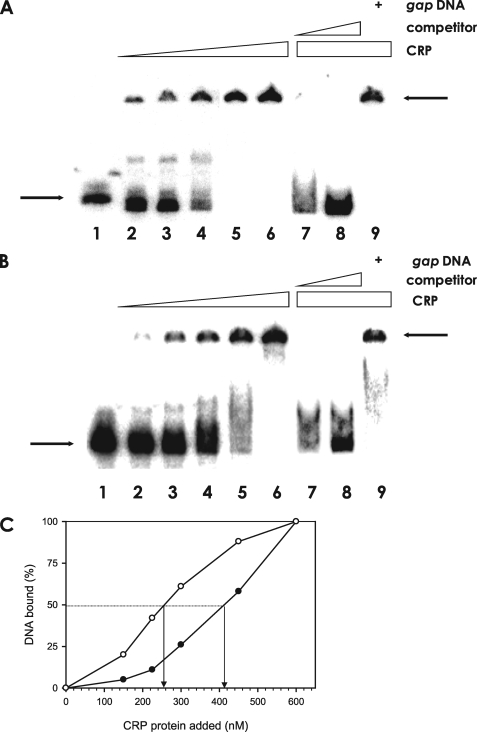
FIGURE 6.
Specific binding of the cAMP-CRP complex to the rpoS promoters. A, binding assay of CRP to the rpoS promoter carrying Pp. The 456-bp DNA fragment of the rpoS upstream region (Pp) was radiolabeled and incubated, with increasing amounts of CRP up to 600 nm. The identical but unlabeled competitor DNA fragment was included in the reaction mixtures in various amounts. Lane 1, probe DNA alone; lanes 2–6, probe DNA incubated with 150, 225, 300, 450, or 600 nm CRP, respectively; lane 7, probe DNA incubated with 600 nm CRP and 14 nm of the identical but unlabeled DNA; lane 8, probe DNA incubated with 600 nm CRP and 70 nm of the identical but unlabeled DNA; and lane 9, probe DNA incubated with 600 nm CRP and 70 nm gap promoter DNA. The arrows on the left side indicate the unbound DNA probe, whereas the arrows on the right side indicate DNA bound to CRP. B, binding assay of CRP to the rpoS promoter carrying Pd. Labeled Pd DNA was incubated with increasing amounts of CRP. For competition analysis, the identical but unlabeled 394-bp DNA fragment was included in the binding reactions containing 600 nm CRP. Lane 1, probe DNA alone; lanes 2–6, probe DNA incubated with 150, 225, 300, 450, or 600 nm CRP, respectively; lane 7, probe DNA incubated with 600 nm CRP and 14 nm of the identical but unlabeled DNA; lane 8, probe DNA incubated with 600 nm CRP and 70 nm of the identical but unlabeled DNA; and lane 9, probe DNA incubated with 600 nm CRP and 70 nm nonspecific gap promoter DNA. The arrows on the left side indicate unbound DNA probe, whereas the arrows on the right side indicate the DNA bound to CRP. C, plot showing the affinity of cAMP-CRP to each promoter of rpoS. The intensities of bound DNA fragments, Pp (open circle) and Pd (closed circle), were estimated by densitometer and plotted against the CRP concentrations. Arrows indicate the concentrations of CRP causing half-maximal binding corresponding to the Kd.