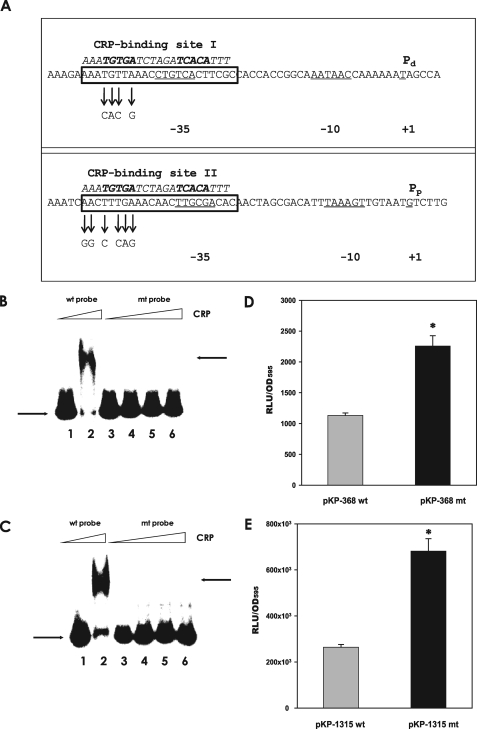
FIGURE 7.
Effect of mutation of the putative CRP-binding sites on CRP binding and rpoS regulation. A, presence of the putative CRP-binding sites in each promoter region of the rpoS gene. Two promoters for the rpoS gene are indicated as Pp and Pd along with their –10 and –35 regions (CRP-binding site I in the Pd promoter and CRP-binding site II in the Pp promoter). Putative CRP-binding sites are presented in the boxes implied from the conserved nucleotide sequences for CRP binding indicated by italicized letters above the V. vulnificus DNA sequence. Site-directed mutagenized nucleotides in the mutant rpoS promoters are indicated with arrows. B, binding assay of CRP to the rpoS promoter carrying Pd and mutagenized Pd. Labeled Pd (wild type probe) and mutagenized Pd DNA (mt probe) were incubated with CRP, as described in Fig. 5. Lane 1, Pd DNA without CRP; lane 2, Pd with 340 nm CRP; lane 3, mutagenized Pd without CRP; lanes 4–6, mutagenized Pd with 340, 510, and 765 nm CRP, respectively. The arrows on the left side indicate the unbound DNA probe, whereas the arrows on the right side indicate the DNA bound to CRP. C, binding assay of CRP to the rpoS promoter carrying Pp and mutagenized Pp. Labeled Pp (wild type probe) and mutagenized Pp DNA (mt probe) were incubated with increasing amounts of CRP. Lane 1, Pp DNA without CRP; lane 2, Pp with 340 nm CRP; lane 3, mutagenized Pp without CRP; lanes 4–6, mutagenized Pp with 340, 510, and 765 nm, CRP, respectively. The arrows on the left side indicate unbound DNA probe, whereas the arrows on the right side indicate the DNA bound to CRP. D and E, effect of mutation on the expression of rpoS::luxAB transcriptional fusions. Wild type V. vulnificus carrying pKP-368 or pKP-368mt (pKP-368 including the mutated CRP-binding site II) and wild type carrying pKP-1315 or pKP-1315mt (pKP-1315, including the mutated CRP-binding site I) were grown in LBS medium supplemented with 3μg/ml tetracycline, and their luciferase activities were measured. Luciferase activities are expressed as normalized values: the number of RLU divided by the A595 value of each sample. Data with p values of <0.01 are indicated with an asterisk.