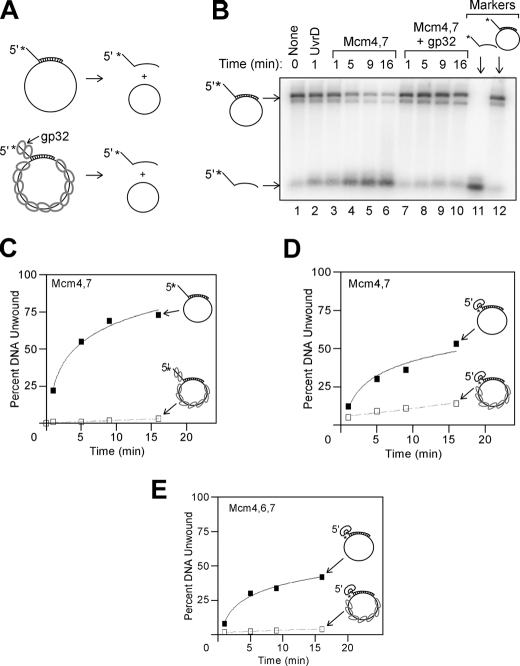
FIGURE 6.
Mcm4/Mcm7 and Mcm4/Mcm6/Mcm7 helicase assemblies can load onto circular ssDNA. A, schematic for experiments to determine whether Mcm assemblies can load onto circular ssDNA. A radiolabeled DNA oligonucleotide is annealed to M13 circular ssDNA. The radiolabeled oligonucleotide bears a 5′-single strand extension to promote steric exclusion, and unwinding of the radiolabeled strand is monitored by native gel electrophoresis. The experiment is also performed in the presence of T4 gp32 to coat the free ssDNA with a nonspecific protein. B, equimolar mixture of Mcm4/Mcm7 was incubated with DNA substrate for the time points incubated at 37 °C in the absence (lanes 3-6) and presence (lanes 7-10) of T4 gp32. The position of free ssDNA and DNA annealed to M13 were determined by standards analyzed on the same gel (lanes 11 and 12). UvrD was also tested (lane 2), as well as a no protein control (lane 1). C, data from experiments similar to B were quantified and plotted as a function of time in the absence (filled squares) and presence (open squares) of T4 gp32. The data in the absence of T4 gp32 were fit to a logarithmic equation, and the data in the presence of T4 gp32 were fit to a linear equation. D, experiment is similar to that shown in C, except the radiolabeled oligonucleotide bears a 5′-biotin-streptavidin in the place of the ssDNA extension. Unwinding rates were determined in the absence (filled squares) and presence (open squares) of T4 gp32. E, experiment is similar to that shown in D, except Mcm4/Mcm6/Mcm7 replaced Mcm4/Mcm7.