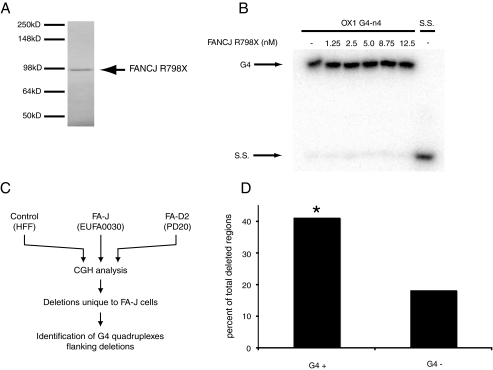
FIGURE 6.
FA-J cells accumulate deletions in regions containing G4 DNA. A, mutant FANCJR798X protein was expressed and purified from Sf9 cells. Protein was resolved by SDS-PAGE and visualized using simply blue stain (Invitrogen). B, FANCJR798X does not unwind G4 DNA substrate in vitro. 32P-End-labeled OX1 G4 DNA was incubated with increasing concentrations of FANCJ (as indicated) in the presence of ATP. Migration of the expected single-stranded oligonucleotide DNA product is indicated (S.S.). Reaction products were analyzed by polyacrylamide gel electrophoresis and visualized using a PhosphorImager (GE Healthcare). C, schematic of array CGH analysis of control, FA-J, and FA-D2 cell lines and subsequent analysis of FA-J-specific deletion breakpoint regions for sequences predicted to form G4 quadruplexes. D, G4 quadruplex DNA flanks deletions in FA-J cells, in an orientation specific manner. 6 kb of sequence up- and downstream of each deletion observed by CGH as specific to FA-J cells was analyzed using the Quadparser algorithm to identify putative G4 DNA signatures. Deletions that associate with G4 DNA signatures were then classified by strand orientation. G4+ designates G4 DNA signatures that lies 3′ to a deleted region, and G4– are those regions that are associated with a G4 DNA signatures situated 5′ to the deleted region. Statistical analysis was performed using a Hidden Markov model to show significance (*, p = 0.04) for the correlation between G4+ DNA signatures and FA-J-specific deleted regions.