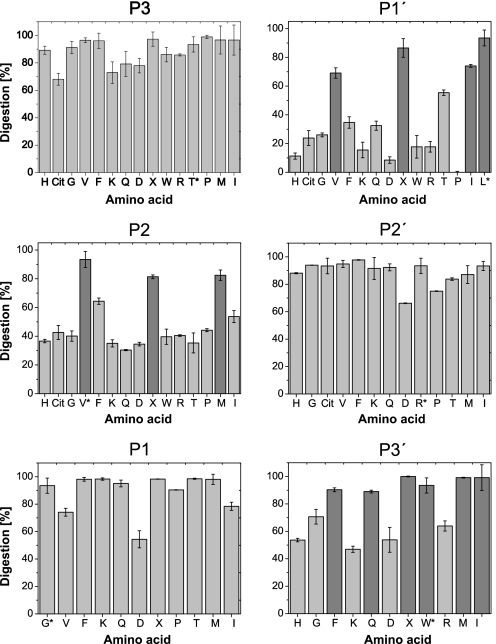
FIGURE 1.
Analysis of CatS specificity. Library peptides with single amino acid exchanges and the general sequence GRWHTVGLRWE-Lys(Dnp)-DArg-NH2 were processed at a concentration of 20 μm for 30 min by 1.3 nm CatS. The reaction mixtures were analyzed by HPLC, and UV peak areas were used for quantification of substrate digestion. All of the assays were performed in triplicate. The error bars denote the means ± standard deviation. The percentage of substrate digestion is shown for peptides with P-3, P-2, P-1, P-1′, P-2′, and P-3′ amino acid modifications. The x axis represents the amino acid at the corresponding substrate position, designated by the single-letter code (X, norleucine; Cit, citrulline). The basis peptide of the library experiment is marked with an asterisk. Preferred amino acids at each substrate position are marked dark gray.