Abstract
The Mur ligases play an essential role in the biosynthesis of bacterial cell-wall peptidoglycan and thus represent attractive targets for the design of novel antibacterials. These enzymes catalyze the stepwise formation of the peptide moiety of the peptidoglycan disaccharide peptide monomer unit. MurC is responsible of the addition of the first residue (l-alanine) onto the nucleotide precursor UDP-MurNAc. Phosphorylation of proteins by Ser/Thr protein kinases has recently emerged as a major physiological mechanism of regulation in prokaryotes. Herein, the hypothesis of a phosphorylation-dependent mechanism of regulation of the MurC activity was investigated in Corynebacterium glutamicum. We showed that MurC was phosphorylated in vitro by the PknA protein kinase. An analysis of the phosphoamino acid content indicated that phosphorylation exclusively occurred on threonine residues. Six phosphoacceptor residues were identified by mass spectrometry analysis, and we confirmed that mutagenesis to alanine residues totally abolished PknA-dependent phosphorylation of MurC. In vitro and in vivo ligase activity assays showed that the catalytic activity of MurC was impaired following mutation of these threonine residues. Further in vitro assays revealed that the activity of the MurC-phosphorylated isoform was severely decreased compared with the non-phosphorylated protein. To our knowledge, this is the first demonstration of a MurC ligase phosphorylation in vitro. The finding that phosphorylation is correlated with a decrease in MurC enzymatic activity could have significant consequences in the regulation of peptidoglycan biosynthesis.
Due to the increasing number of antibiotic-resistant strains and the emergence of new pathogenic microorganisms, one of the biggest challenges for modern biomedical research is the continuous development of new antimicrobial drugs targeting bacterial essential mechanisms such as cell division or peptidoglycan (PG)4 biosynthesis (1). The bacterial cell wall PG is a giant molecule that sustains the shape of the bacterial cell and contains the outward forces generated in maintaining an osmotic pressure gradient against the environment. Without this PG layer the cell integrity would be ruptured, and this could lead to cell death. Therefore the PG biosynthesis machinery represents a promising source of putative targets for antibacterial chemotherapy (2, 3).
The biosynthesis of bacterial PG is a complex two-stage process (4). The first stage involves the assembly of the disaccharide peptide monomer unit by enzymes located in the cytoplasm or at the inner surface of the cytoplasmic membrane (3, 5). The peptide moiety of the monomer unit is assembled stepwise by the successive additions of l-alanine, d-glutamic acid, meso-diaminopimelic acid or l-lysine, and d-alanyl-d-alanine to UDP-N-acetylmuramic acid (UDP-MurNAc). These steps are catalyzed by specific peptide synthetases (ligases), which are designated as MurC, MurD, MurE, and MurF, respectively, all participating in non-ribosomal peptide bond formation with the concomitant hydrolysis of ATP. The MurNAc-pentapeptide motif of the resulting nucleotide precursor is then transferred by the MraY translocase onto the undecaprenyl phosphate carrier molecule, generating the lipid intermediate I. The subsequent addition of the N-acetylglucosamine motif of UDP-GlcNAc onto lipid I generates lipid II in a reaction catalyzed by MurG (6). The second stage of the PG biosynthesis consists in the polymerization by transglycosylation and transpeptidation reactions of the disaccharide pentapeptide monomers, a reaction taking place in the periplasmic space and that is catalyzed by the penicillin-binding proteins.
The PG biosynthesis pathway enzymes, which are essential and specific for bacteria, represent important potential targets for screening novel antibacterial compounds. Due to the growing emergence of bacterial multiresistance to currently used antibiotics, the discovery of new therapeutic compounds has indeed become a necessity. In recent years, an extensive search for specific inhibitors interfering with the cytoplasmic steps of this pathway, and in particular with the four steps catalyzed by the Mur ligases, has been developed (2, 7, 8). The UDP-Mur-NAc:l-alanine ligase (MurC), encoded by the murC gene, represents such an interesting candidate for drug development (9, 10). Recently, different phosphinic acid derivatives and substrate analogues have been identified as Mur ligase inhibitors (11, 12).
Corynebacterium glutamicum is a rod-shaped non-pathogenic Gram-positive actinomycete widely used in the industrial production of amino acids such as l-lysine and l-glutamic acid (13). C. glutamicum has been extensively studied as a model microorganism due to the strategies employed by this actinomycete to achieve a rod-shaped morphology. In fact, the mechanisms taking place in C. glutamicum happened to be completely different from that of Escherichia coli or Bacillus subtilis (14, 15), whereas the number of genes involved in cell division and PG biosynthesis in C. glutamicum is lower (16).
Interestingly, an earlier work on the phosphoproteome of C. glutamicum (17) identified MurC has being phosphorylated in vivo, suggesting that protein phosphorylation plays a much broader function in C. glutamicum than was previously expected. Recently, we described the characterization of the four STPKs from C. glutamicum ATCC 13869 and highlighted their role in cell division (18). Moreover, Thakur and Chakraborti (19) showed that MurD from Mycobacterium tuberculosis was phosphorylated by the Ser/Thr protein kinase (STPK) PknA, although no further characterization of the role of the phosphorylation on the MurD enzyme activity was investigated. Therefore, it was tempting to speculate that MurC in C. glutamicum could also be regulated by STPK phosphorylation.
The focus of this work is to study the regulation of MurC in C. glutamicum via phosphorylation. As a first step in deciphering the potential role/participation of the corynebacterial STPKs in the regulation of MurC activity, we confirmed its specific phosphorylation by the PknA kinase through a combination of in vitro phosphorylation assays and mass spectrometric identification of the different MurC phosphorylation sites. Moreover, we demonstrated that the murein ligase activity of MurC was negatively regulated upon its phosphorylation. To our knowledge, this work represents the first evidence of a Mur enzyme regulated by phosphorylation.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions—Bacterial strains and plasmids are described in Table 1. Strains used for cloning and expression of recombinant proteins were E. coli TOP10 (Invitrogen) and E. coli BL21(DE3)Star (Stratagene), respectively. E. coli cells were grown and maintained at 37 °C in LB medium supplemented with 100 μg/ml ampicillin and/or 50 μg/ml kanamycin, when required. The murC temperature-sensitive E. coli strain H1119 was grown at 30 °C in 2YT (1.6% Bactotrypton, 1.0% Bactoyeast extract, 0.5% NaCl, pH 7.0) medium and was used for genetic complementation experiments with plasmids carrying wild-type or mutated copies of the murC gene. C. glutamicum cells were grown at 30 °C in TSB (Trypticase soy broth, Oxoid) or TSA (TSB containing 2% agar) medium supplemented with 12.5 μg/ml kanamycin. Plasmids to be transferred by conjugation from E. coli to corynebacteria were introduced by transformation into the donor strain E. coli S17-1. Mobilization of plasmids from E. coli S17-1 to C. glutamicum R31 was accomplished as described previously (20).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Genotype or description | Source or reference |
|---|---|---|
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG; used for general cloning | Invitrogen |
| E. coli BL21 (DE3) Star | F2 ompT hsdSB(rB2 mB2) gal dcm (DE3); used to express recombinant proteins in E. coli | Stratagene |
| E. coli H1119 | murC temperature-sensitive mutant | (23) |
| E. coli S17-1 | Mobilizing donor strain, pro recA, which possesses an RP4 derivative integrated into the chromosome | (38) |
| C. glutamicum 13869 | Wild-type control strain | ATCC |
| C. glutamicum R31 | C. glutamicum ATCC 13869; derivative used as recipient in conjugation experiments | (39) |
| pETTev | pET15b (Novagen) derivative including the replacement of the thrombin site coding sequence with a tobacco etch virus (TEV) protease site | (40) |
| pTEVmurC | pTEV derivative used to express His-tagged fusion of MurC | This work |
| pGEX4T-3 | E. coli vector designed to make GST gene fusions | GE Healthcare |
| pGEXA | pGEX4T-3 derivative used to express GST fusion of PknA cytoplasmic domain | (18) |
| pGEXB | pGEX4T-3 derivative used to express GST fusion of PknB cytoplasmic domain | (18) |
| pGEXL | pGEX4T-3 derivative used to express GST fusion of PknL cytoplasmic domain | (18) |
| pTEVGfull | pETTev derivative used to express His-tagged PknG | (18) |
| pEDiv | Mobilizable plasmid able to replicate in E. coli and C. glutamicum; kan and cat resistance genes | A. Ramos (unpublished) |
| pEDivmurHis | pEDiv derivative used to express His-tagged fusion of MurC in C. glutamicum | This work |
| pGEXpknAT179A | pGEXA derivative used to express GST fusion of PknA cytoplasmic domain carrying the mutation T179A | This work |
| pGEXpknAT181A | pGEXA derivative used to express GST fusion of PknA cytoplasmic domain carrying the mutation T181A | This work |
| pGEXpknAT179/181A | pGEXA derivative used to express GST fusion of PknA cytoplasmic domain carrying the mutations T179A and T181A | This work |
| pGEXpknAK49A | pGEXA derivative used to express GST fusion of PknA cytoplasmic domain carrying the mutation K49A | This work |
| pTEVmurC1T | pTEVmurC derivative used to express His-tagged fusion of MurC1T carrying the mutation T362A | This work |
| pTEVmurC2T | pTEVmurC1T derivative used to express His-tagged fusion of MurC2T carrying the mutation T362A/T365A | This work |
| pTEVmurC3T | pTEVmurC2T derivative used to express His-tagged fusion of MurC3T carrying the mutation T362A/T365A/T51A | This work |
| pTEVmurC4T | pTEVmurC3T derivative used to express His-tagged fusion of MurC4T carrying the mutation T362A/T365A/T51A/T120A | This work |
| pTEVmurC5T | pTEVmurC4T derivative used to express His-tagged fusion of MurC5T carrying the mutation T362A/T365A/T51A/T120A/T167A | This work |
| pTEVmurC6T | pTEVmurC5T derivative used to express His-tagged fusion of MurC6T carrying the mutation T362A/T365A/T51A/T120A/T167A/T133A | This work |
| pTEVmurCT51A | pTEVmurC derivative used to express His-tagged fusion of MurCT51A carrying the mutation T51A | This work |
| pTEVmurCT120A | pTEVmurC derivative used to express His-tagged fusion of MurCT120A carrying the mutation T120A | This work |
| pTEVmurCT133A | pTEVmurC derivative used to express His-tagged fusion of MurCT133A carrying the mutation T133A | This work |
| pTEVmurCT167A | pTEVmurC derivative used to express His-tagged fusion of MurCT167A carrying the mutation T167A | This work |
| pTEVmurCT362A | pTEVmurC derivative used to express His-tagged fusion of MurCT362A carrying the mutation T362A | This work |
| pTEVmurCT365A | pTEVmurC derivative used to express His-tagged fusion of MurCT365A carrying the mutation T365A | This work |
| pTrc99A | E. coli vector allowing high level expression under the IPTG inducible trc promoter | Pharmacia |
| pTrc99murC | pTrc99A derivative carrying murC | This work |
| pTrc99murC1T | pTrc99A derivative carrying murC1T (T362A) | This work |
| pTrc99murC2T | pTrc99A derivative carrying murC2T (T362A/T365A) | This work |
| pTrc99murC3T | pTrc99A derivative carrying murC3T (T362A/T365A/T51A) | This work |
| pTrc99murC4T | pTrc99A derivative carrying murC4T (T362A/T365A/T51A/T120A) | This work |
| pTrc99murC5T | pTrc99A derivative carrying murC5T (T362A/T365A/T51A/T120A/T167A) | This work |
| pTrc99murC6T | pTrc99A derivative carrying murC6T (T362A/T365A/T51A/T120A/T167A/T133A) | This work |
Cloning, Expression, and Purification of MurC Proteins—First, the murC gene was cloned to generate a recombinant MurC protein expressed in E. coli. Therefore, the murC gene was amplified by PCR using C. glutamicum ATCC 13869 genomic DNA as a template and the primers pair murC1/murC2 (Table 2), containing NdeI and NheI restriction sites, respectively. The 1461-bp amplified product was digested by NdeI and NheI and ligated to the pETTev vector (Table 1) generating the pTEVmurC plasmid. E. coli BL21(DE3)Star cells transformed with this construction were used for expression and purification of His6-tagged MurC, as previously described (21). Finally, the purified His6-tagged MurC was treated with TEV protease according to the manufacturer's instructions (Invitrogen). Secondly, overexpression and purification of MurC from C. glutamicum cultures was performed using standard PCR strategies. The murC gene from C. glutamicum was amplified using the primers pair murC1/murHisNdeI2 (Table 2). The PCR product carrying a His tag at its C-terminal end was digested with NdeI and subsequently cloned under the control of the Pdiv promoter into plasmid pEDiv (Table 1). The resulting expression vector, named pEDivmurHis, was introduced by conjugation into C. glutamicum R31. Purification of the soluble His6-tagged MurC protein from C. glutamicum was performed as described previously (21).
TABLE 2.
Primers used in this study
| Primer | Gene | 5′ to 3′ Sequenceab |
|---|---|---|
| murC1 | murC | GGAATTCCATATGGTGACCACTCCACAC (NdeI) |
| murC2 | murC | TGGAATTCGCTAGCCTAATTGTTTTGCAGCTGATCC (NheI) |
| murHisNdeI | murC | GGAATTCCATATGCTAATGATGATGATGATGATGATTGTTTTGCAGC (NdeI) |
| N-pknAK49M | pknA | GATCGCGAAGTAGCCATCATGGTACTGCGCCCGGAATTTTCC |
| C-pknAK49M | pknA | GGAAAATTCCGGGCGCAGTACCATGATGGCTACTTCGCGATC |
| N-pknAT179A | pknA | GCCGCTGCTGTGCCTTTGGCCCGCACCGGCATGGTGGTG |
| C-pknAT179A | pknA | CACCACCATGCCGGTGCGGGCCAAAGGCACAGCAGCGGC |
| N-pknAT181A | pknA | GCTGTGCCTTTGACCCGCGCCGGCATGGTGGTGGGTACT |
| C-pknAT181A | pknA | AGTACCCACCACCATGCCGGCGCGGGTCAAAGGCACAGC |
| N-pknAT179A/T181A | pknA | GCCGCTGCTGTGCCTTTGGCCCGCGCCGGCATGGTGGTGGGT |
| C-pknAT179A/T181A | pknA | ACCCACCACCATGCCGGCGCGGGCCAAAGGCACAGCAGCGGC |
| N-murC362 | murC | GATTACGCACACCACCCAGCGGAAGTAACTGCAGTGCTC |
| C-murC362 | murC | GAGCACTGCAGTTACTTCCGCTGGGTGGTGTGCGTAATC |
| N-murC362/365 | murC | GCACACCACCCAGCGGAAGTAGCTGCAGTGCTCAGCGCTGCG |
| C-murC362/365 | murC | CGCAGCGCTGAGCACTGCAGCTACTTCCGCTGGGTGGTGTGC |
| N-murC365 | murC | CACCACCCAACGGAAGTAGCTGCAGTGCTCAGCGCGGCG |
| C-murC365 | murC | CGCAGCGCTGAGCACTGCAGCTACTTCCGTTGGGTGGTG |
| N-murC51 | murC | GATGCCAAAGATTCCCGCGCCTTGCTTCCACTCCGCGCC |
| C-murC51 | murC | GGCGCGGAGTGGAAGCAAGGCGCGGGAATCTTTGGCATC |
| N-murC120 | murC | GAATTGCTGGAAGGCTCCGCCCAGGTCTTGATCGCGGGT |
| C-murC120 | murC | ACCCGCGATCAAGACCTGGGCGGAGCCTTCCAGCAATTC |
| N-murC167 | murC | ACCAATGCGCACCATGGAGCTGGTGAGGTCTTTATCGCT |
| C-murC167 | murC | AGCGATAAAGACCTCACCAGCTCCATGGTGCGCATTGGT |
| N-murC133 | murC | ACCCACGGTAAGACCTCCGCCACCTCTATGTCTGTGGTA |
| C-murC133 | murC | TACCACAGACATAGAGGTGGCGGAGGTCTTACCGTGGGT |
| murH1119-1 | murC | TTTAATCATGACCACTCCACACTTGG (BspHI) |
| murH1119-2 | murC | CTTACAGATCTCTAATTGTTTTGCAGCTG (BglII) |
Restriction sites are underlined and specified by brackets.
Mutagenized codons are shown in bold.
In Vitro Kinase Assays—In vitro phosphorylation was performed with 2 μg of MurC in 20 μl of buffer P (25 mm Tris-HCl, pH 7.0, 1 mm dithiothreitol, 5 mm MgCl2, 1 mm EDTA) with 200 μCi/ml [γ-33P]ATP corresponding to 65 nm (PerkinElmer Life Sciences, 3000 Ci/mmol), and 0.5 μg of kinase. Plasmids pGEXA, pGEXB, pGEXL, and pTEVGfull (Table 1) were used for the expression and purification in E. coli of the four recombinant STPKs from C. glutamicum as previously described (18). After 15-min incubation, the reaction was stopped by adding sample buffer and heating the mixture at 100 °C for 5 min. The reaction mixtures were analyzed by SDS-PAGE. After electrophoresis, gels were soaked in 20% trichloroacetic acid for 10 min at 90 °C, stained with Coomassie Blue, and dried. Radioactive proteins were visualized by autoradiography using direct exposure films.
Analysis of the Phosphoamino Acid Content of Proteins—MurC sample (5 μg) phosphorylated in vitro by the GST-tagged PknA, and unreacted [γ-33P]ATP were separated by one-dimensional gel electrophoresis and electroblotted onto an Immobilon polyvinylidene difluoride membrane. The 33P-labeled protein bands were detected by autoradiography and excised from the Immobilon blot and hydrolyzed in 6 m HCl for 1 h at 110 °C. The acid-stable phosphoamino acids released were separated by electrophoresis in the first dimension at pH 1.9 (800 V/h) in 7.8% acetic acid, 2.5% formic acid, followed by ascending chromatography in the second dimension in 2-methyl-1-propanol/formic acid/water (8:3:4, v/v). After migration, radioactive compounds were detected by autoradiography. Authentic phosphoserine, phosphothreonine and phosphotyrosine were run in parallel and visualized by staining with ninhydrin.
Cloning and Purification of PknA Mutant Proteins—Site-directed mutagenesis was directly performed on the pGEXA expression plasmid (Table 1) using inverse-PCR amplification with the following self-complementary primers (Table 2): N-pknAT179A/C-pknAT179A, N-pknAT181A/C-pknAT181A, N-pknAT179A-T181A/C-pknAT179A-T181A, and N-pknAK49M/C-pknAK49M to generate pGEXpknAT179A, pGEXpknAT181A, pGEXpknAT179A/T181A, and pGEXpknAK49M, respectively (Table 1). All constructs were verified by DNA sequencing. The different GST-tagged recombinant fusion proteins were overexpressed and purified as reported earlier (18).
Site-directed Mutagenesis—The six threonine residues from C. glutamicum MurC identified by mass spectrometry after in vitro phosphorylation with GST-tagged PknA were replaced by alanine residues by site-directed mutagenesis using inverse-PCR amplification. A first PCR was carried out using pTEVmurC (Table 1) as a template with the primers pair N-murC362 and C-murC362 (Table 2) to generate pTEVmurC1T (T362A). A second PCR was carried out using pTEVmurC (Table 1) as a template with the primers pair N-murC362/365 and C-murC362/365 (Table 2) to generate pTEVmurC2T (T362A/T365A). This mutant and the subsequent additional mutants were used as templates in subsequent PCR reactions using the following primers pairs: N-murC51 and C-murC51, N-murC120 and C-murC120, N-murC167 and C-murC167, and N-murC133 and C-murC133 (see Table 2) to generate pTEVmurC3T (T362A/T365A/T51A), pTEVmurC4T (T362A/T365A/T51A/T120A), pTEVmurC5T (T362A/T365A/T51A/T120A/T167A), and pTEVmurC6T (T362A/T365A/T51A/T120A/T167A/T133A), respectively. Individual mutants were generated using pTEVmurC (Table 1) as a template with the primers pairs N-murC51/C-murC51, N-murC120/C-murC120, N-murC133/C-murC133, N-murC167/C-mur-C167, N-murC362/C-murC362, and N-murC365/C-murC365 (Table 2) to generate pTEVmurCT51A, pTEVmurCT120A, pTEVmurCT133A, pTEVmurCT167A, pTEVmurCT362A, and pTEVmurC T365A, respectively (Table 1). All the resulting constructs were verified by DNA sequencing. The different His6-tagged mutant proteins were overexpressed and purified, as described above.
MS Analysis—Purified wild-type and mutant MurC proteins were subjected to in vitro phosphorylation by GST-tagged PknA as described above, excepted that [γ-33P]ATP was replaced with 5 mm ATP. Subsequent analyses using NanoLC/nanospray/tandem mass spectrometry (LC-ESI/MS/MS) were performed as previously described (18).
Immunoblotting—Corynebacterial MurC purified either from E. coli or C. glutamicum was loaded on a 10% polyacrylamide gel, electrophoresed, blotted on polyvinylidene difluoride, and detected using either monoclonal mouse anti-phospho-threonine, -serine, -tyrosine, or polyclonal rabbit anti-His antibodies used at 1:100 and 1:10,000 dilution, respectively. Alkaline phosphatase-conjugated anti-mouse or anti-rabbit was used as a secondary antibody at a 1:5,000 dilution.
Complementation with C. glutamicum MurC—Plasmids pTEVmurC, pTEVmurC1T, pTEVmurC2T, pTEVmurC3T, pTEVmurC4T, pTEVmurC5T, and pTEVmurC6T were used as templates for PCR amplification of the murC gene with the primers pair murH1119-1/murH1119-2 (Table 2) containing a BspHI and BglII restriction site, respectively. The resulting 1461-bp products, carrying the wild-type and mutated gene copies, respectively, were digested with BspHI and BglII, and cloned into the plasmid vector pTrc99A (22) between the compatible NcoI and BamHI sites, generating pTrcmurC, pTrcmurC1T, pTrcmurC2T, pTrcmurC3T, pTrcmurC4T, pTrcmurC5T, and pTrcmurC6T, respectively. These plasmids allowing high level expression of the C. glutamicum MurC and MurC mutants under the control of the strong isopropyl 1-thio-β-d-galactopyranoside-inducible trc promoter, were transformed in the E. coli MurC temperature-sensitive mutant strain H1119 (23). Transformants were grown at the permissive temperature (30 °C) before being shifted to the restrictive temperature (42 °C). Complementation was judged by the ability of the mutant to grow at the restrictive temperature.
UDP-MurNAc l-alanine Ligase Assay—The l-alanine-adding activity of wild-type and mutant MurC proteins was assayed according to Liger et al. (24) by following the formation of UDP-MurNAc-l-[14C]alanine in 40 μl of reaction mixture containing 100 mm Tris-HCl, pH 8.6, 20 mm MgCl2, 20 mm ammonium sulfate, 0.5 mm l-[14C]alanine (0.6 KBq, 5.5 Gbq/mmol, Amersham), 1 mm UDP-MurNAc, 5 mm ATP, and enzyme (25 μl of an appropriate dilution in buffer A: 20 mm potassium phosphate, pH 7.2, containing 1 mm dithiothreitol and 10% glycerol). The mixture was incubated at 37 °C for 30 min and the reaction was stopped by addition of 8 μl of acetic acid, followed by lyophilization. To test the effect of the phosphorylation of MurC on its enzyme activity, the assay was performed as described above excepted that the MurC enzyme (20 μg/ml) was preincubated overnight at 37 °C with or without wild-type PknA (16 μg/ml), or with PknA_K49M mutant (30 μg/ml) in buffer A supplemented with 2 mm ATP and 5 mm MgCl2. The radioactive product UDP-MurNAc-l-[14C]alanine and substrate l-[14C]alanine were separated by reversed-phase high-performance liquid chromatography on a Nucleosil 5C18 column (4.6 × 150 mm, Alltech France) using 50 mm ammonium formate, pH 3.9, as the eluent, at a flow rate of 0.6 ml/min. Detection was performed with a radioactive flow detector (model LB506-C1, Berthold France, La Garenne-Colombes, France) using the Quicksafe Flow 2 scintillator (Zinsser Analytic, Maidenhead, UK) at 0.6 ml/min. Quantitation was carried out with the Radiostar software (Berthold).
RESULTS AND DISCUSSION
MurC Is a Substrate of the C. glutamicum PknA—The peptide moiety of the PG monomer unit is assembled stepwise in the cytoplasm by the successive actions of four Mur ligases designated as MurC, MurD, MurE, and MurF. These enzymes share limited sequence identity but have several highly conserved regions that map primarily to the active site. Each of them comprises three structural domains: an N-terminal domain with a Rossmann-type fold primarily responsible for binding of the UDP-MurNAc(-peptide) substrate, a large central ATP-binding (ATPase) domain, and a C-terminal domain associated with binding of the amino acid substrate (25). Based on protein sequence alignments and on the three-dimensional structure of the E. coli MurC protein (26), these three domains in the C. glutamicum MurC may extend between Met1-Gly118, Ser119-Arg334, and Arg335-Asn486, respectively (Figs. 1 and 4). By comparing the amino acid sequences of the MurC, -D, -E, and -F ligases from different bacterial genera, several conserved residues were identified. This analysis led to the characterization of the ATP-binding motif located in the conserved sequence (GXXGK(T/S)), and therefore named the GKT motif (Fig. 1). There are also a number of common conserved amino acids residues present in the different Mur ligases. Taking E. coli as a model, it was established that Asp50, Lys130, Glu174, and Asp351 were essential for the catalytic process of these ligases, whereas residues His199, Asn293, Asn296, and Arg327 were involved in the structure of the active site. The sequence of several MurC proteins from representative species of mycobacteria and corynebacteria were aligned using the ClustalW and Espript programs (Fig. 1). The invariant residues Asp50, Lys130, Glu174, Asn296, and Arg327 appeared conserved in all the sequences aligned, thus confirming that MurC from C. glutamicum harbors most of the characteristics of a functional Mur ligase enzyme. Therefore, the recombinant MurC protein was expressed and purified from E. coli BL21(DE3)Star harboring the pTEVmurC. The protein contained an N-terminal His tag, which was subsequently removed after cleavage with the TEV protease. Analysis of the purified recombinant protein by SDS-PAGE revealed that the protein was expressed in a soluble form migrating slightly above its predicted molecular mass of 52 kDa (Fig. 2A).
FIGURE 1.
Multiple sequence alignment of MurC ortholog proteins from corynebacteria, mycobacteria, and E. coli. The alignment was performed using ClustalW and Espript programs (C_glu, C. glutamicum; C_eff, C. efficiens; C_dip, C. diphtheriae; C_jei, C. jeikeium; M_tub, M. tuberculosis; M_sme, M. smegmatis; and E_col, E. coli). The conserved GKT motif involved in ATP binding is represented by a shaded oval. Triangles represent PknA-dependent phosphorylation sites (Thr51, Thr120, Thr133, Thr167, Thr362, and Thr365) of C. glutamicum MurC. Residues indicated by an asterisk in the E. coli MurC sequence correspond to residues that were previously identified as invariant within the whole Mur ligase family (3, 25): the Asp50, Lys130 (GKT motif), and Glu174 residues involved in the catalytic process, and the Asn296 and Arg327 residues involved in the structural organization of the active site. Numbering of amino acids corresponds to the MurC protein from C. glutamicum.
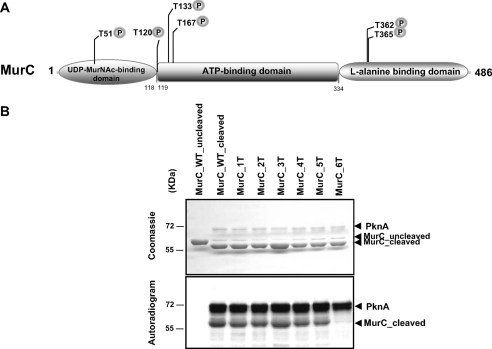
FIGURE 4.
Structural organization of MurC and phosphorylation of MurC mutants by PknA. A, schematic representation of MurC from C. glutamicum. The MurC protein comprises three structural domains, an N-terminal domain responsible for the binding of the UDP-MurNAc substrate, a central ATP-binding (ATPase) domain, and a C-terminal domain involved in the binding of the amino acid substrate l-alanine. These domains are shown by shaded circles and boxes. The phosphorylation sites identified in MurC are indicated by a “P.” B, in vitro phosphorylation of MurC mutants by PknA. The different MurC mutants were treated with the TEV protease to remove the N-terminal His tag and then used in phosphorylation assays in equal amounts in the presence of [γ-33P]ATP and PknA. The MurC_1T, MurC_2T, MurC_3T, MurC_4T, MurC_5T, and MurC_6T mutant proteins corresponding to MurC_T362A, MurC_T362A/T365A, MurC_T362A/T365A/T51A, MurC_T362A/T365A/T51A/T120A, MurC_T362A/T365A/T51A/T120A/T167A, and MurC_T362A/T365A/T51A/T120A/T167A/T133A, respectively, were separated by SDS-PAGE and stained with Coomassie Blue (upper panel), and the radioactive bands were revealed by autoradiography (lower panel).
FIGURE 2.
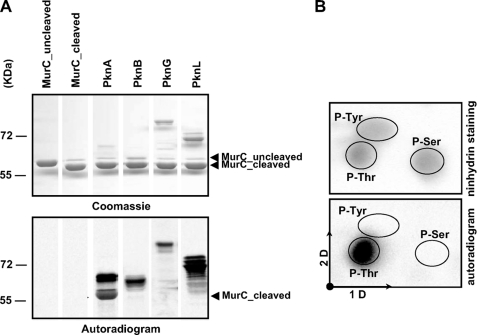
In vitro phosphorylation of C. glutamicum MurC. A, in vitro phosphorylation of MurC by corynebacterial STPKs. The four recombinant STPKs (PknA, PknB, PknG, and PknL) were expressed and purified as previously described by Fiuza et al. (18). Recombinant MurC was treated with the TEV protease to remove the N-terminal His tag and then incubated with [γ-33P]ATP and the different kinases. Samples were separated by SDS-PAGE (upper panel) and visualized by autoradiography (lower panel). Upper bands illustrate the autokinase activity of each STPK, whereas lower bands reflect phosphorylation of MurC. B, phosphoamino acid content of MurC. MurC was phosphorylated in vitro in presence of PknA and [γ-33P]ATP, analyzed by SDS-PAGE, electroblotted onto an Immobilon polyvinylidene difluoride membrane, excised, and hydrolyzed in acid. The phosphoamino acids thus liberated were separated by electrophoresis in the first dimension (1D) and ascending chromatography in the second dimension (2D). After migration, radioactive molecules were detected by autoradiography (lower panel). Authentic phosphoserine (P-Ser), phosphothreonine (P-Thr), and phosphotyrosine (P-Tyr) were run in parallel as internal standard controls, and visualized by ninhydrin staining (upper panel).
Previous work on the phosphoproteome of C. glutamicum (17) identified MurC as being phosphorylated in vivo. Moreover, we have recently reported the characterization of the four STPKs from C. glutamicum ATCC 13869 and highlighted their role in cell division (18). These results prompted us to investigate whether MurC would also represent a substrate for corynebacterial STPKs. A systematic approach was used to investigate whether STPKs of C. glutamicum (PknA, PknB, PknG, or PknL) could phosphorylate MurC. All these STPKs were expressed and purified from E. coli as described previously (18). The different STPKs migrate as diffuse bands reflecting the different levels of phosphorylation for each isoform, and this aberrant profile of migration of STPKs kinases has already been reported in earlier studies (27-29). Interestingly, when STPKs were incubated in the presence of recombinant MurC and [γ-33P]ATP, phosphorylation of MurC was specifically observed with the PknA kinase, whereas PknB and PknL, which display autophosphorylation activity in vitro, did not phosphorylate MurC (Fig. 2A). The PknG kinase needs to be activated by the PknA kinase to trigger its autophosphorylation activity, therefore PknG was phosphorylated by PknA as previously described and then used to transphosphorylate MurC (18). As shown on Fig. 2A, MurC did not show any radioactive band, thus indicating that PknG, even if activated by its cognate kinase PknA, could not phosphorylate MurC in vitro. Moreover, MurC alone is unable to incorporate [γ-33P]ATP, confirming that MurC is a substrate of PknA (Fig. 2A).
We next determined the nature of the amino acid residues phosphorylated in MurC by analyzing the phosphoamino acid content of PknA-phosphorylated MurC. The MurC protein (5 μg) was labeled with [γ-33P]ATP in vitro, separated by SDS-PAGE, excised, and subjected to acid hydrolysis. Fig. 2B shows that MurC was exclusively phosphorylated on threonine residues.
Together, these data suggest a specificity of substrate recognition by PknA toward MurC. Although a specific interaction between MurC and PknA may exist, it remains to be established whether this specific partnering also occurs in vivo.
The Activation Loop Thr179 and Thr181 Residues Are Essential for the Autophosphorylation Activity of C. glutamicum PknA—Diverse mechanisms of eukaryotic protein kinase regulation have been described. The transition between active and inactive forms may occur via control of access to the catalytic site and/or the substrate-binding site. These regulatory mechanisms involve phosphorylation/dephosphorylation via an autocatalytic mechanism or the action of other kinases and phosphatases. The activation loop appears to be a major control element of an active/inactive conformational switch in numerous kinases (30), and the conformation of the activation loop often depends on the phosphorylation state (31). Based on structural studies, it is thought that the activation loop controls both the accessibility to the catalytic site and the binding of the substrate. Recently, Canova et al. (27) demonstrated that the phosphorylated residues Thr173 and Thr175 present in the activation loop were essential for the autophosphorylation activity of the M. tuberculosis PknL kinase, and that phosphorylation of the Thr173 residue was also required for optimal PknL-mediated phosphorylation of its substrate Rv2175c. Interestingly, the corresponding threonine residues Thr179 and Thr181 found in the C. glutamicum PknA were recently identified as phosphorylation sites (18). The presence of such residues in PknA from C. glutamicum supports the concept that phosphorylation of the activation loop could play a regulatory role, as recently described for other mycobacterial STPKs (27, 32, 33).
To determine a possible link between Thr179 and Thr181 and the autophosphorylation activity of PknA, the two residues were individually substituted by Ala to generate PknA_T179A and PknA_T181A, respectively. In addition, a double mutant was also created and designated PknA_T179A/T181A. These PknA mutants were expressed and purified from E. coli as previously described (18). The electrophoretic migration profile showed that the different PknA mutants migrated differently from the wild-type protein (Fig. 3A, upper panel). We reasoned that, because these proteins displayed different electrophoretic mobility properties, they must differ in their intrinsic phosphorylation states. This hypothesis was confirmed by labeling the mutant proteins in vitro with [γ-33P]ATP. PknA_T179A gave rise to a lower radioactive signal than the wild-type enzyme (Fig. 3A, lower panel). Quantification of the signal intensity indicated an autophosphorylation activity of 47% with respect to the activity of the wild-type protein, which was arbitrarily placed at 100%. This result argues that Thr179 is a key phosphorylation site of the activation loop, necessary for PknA autophosphorylation activity. The signal intensity generated by PknA_T181A was intermediate between those of the wild-type and Thr179 mutant proteins (Fig. 3A, lower panel), representing 81% of residual activity with respect to the wild-type protein activity. Therefore, this result confirms the requirement of Thr181 for optimal PknA autophosphorylation activity, although this residue appears less important than Thr179. When both Thr residues were mutated, the intensity of the signal was similar to the one of the single Thr179 mutant (Fig. 3A, lower panel). Together, these results confirm that double phosphorylation of the activation loop residues Thr179 and Thr181 is necessary for full kinase activity and unambiguously demonstrate the role of both phosphothreonines.
FIGURE 3.
In vitro phosphorylation of MurC by PknA and the PknA activation loop mutants. A, in vitro phosphorylation of the different PknA activation loop mutants. All proteins were overproduced in E. coli and purified as GST fusions. The following proteins were incubated with [γ-33P]ATP: PknA_WT, PknA_K49M, PknA_T179A, PknA_T181A, and PknA_T179A/T181A. Proteins were separated by SDS-PAGE and stained with Coomassie Blue (upper panel), and the radioactive bands were revealed by autoradiography (lower panel). B, in vitro phosphorylation of MurC by PknA and the different PknA activation loop mutants. Recombinant MurC was treated with the TEV protease to remove the N-terminal His tag and then used in phosphorylation assays in the presence of [γ-33P]ATP and PknA_WT, PknA_K49M, PknA_T179A, PknA_T181A, or PknA_T179A/T181A. Proteins were separated by SDS-PAGE and stained with Coomassie Blue (upper panel), and the radioactive bands were revealed by autoradiography (lower panel).
To exclude the possibility of exogenous contamination that could explain labeling of PknA with [γ-33P]ATP, Lys49 was mutated to Met. Purified PknA_K49M, was incubated in the presence of [γ-33P]ATP, and as expected, no signal could be detected as Lys49 is essential for catalyzing the phosphorylation reaction, in agreement with previous reports (Fig. 3A, lower panel) (34, 35). This confirms that the radioactive signals observed for the different isoforms of the PknA kinase are only representing the autophosphorylation activity of PknA and not a contamination from the purification procedure.
Phosphorylation of MurC Is Dependent on the PknA Activation Loop Thr179 Residue—Several recent publications indicated that phosphorylation of the threonine residues present in the STPK activation loop is not only necessary for controlling the kinase activity but is also required for recruitment and phosphorylation of its substrate (27, 36). These results prompted us to analyze the contribution of the activation loop key residues of PknA (Thr179 and Thr181) in the transphosphorylation mechanism between the kinase and its substrate MurC. This was achieved by incubating recombinant MurC with PknA_K49M, PknA_T179A, PknA_T181A, or PknA_T179A/T181A in the presence of [γ-33P]ATP (Fig. 3B). PknA_K49M was unable to phosphorylate C. glutamicum MurC, indicating that phosphorylation of PknA is a prerequisite to the transphosphorylation reaction (Fig. 3B, lower panel). More importantly, whereas the T179A mutation completely abolished the transphosphorylation reaction, replacement of Thr181 by Ala did not alter PknA-dependent phosphorylation of MurC (Fig. 3B, lower panel). Furthermore, MurC could not be phosphorylated by PknA_T179A/T181A, clearly demonstrating that phosphorylation of MurC is dependent on Thr179. The finding that MurC interacts only with the phosphorylated form of PknA and that this interaction is abolished by the T179A substitution suggests that this phosphopeptide recognition motif is involved in protein-protein interaction between the two corynebacterial partners, as previously described in its related actinomycete M. tuberculosis (27).
MurC Is Phosphorylated on Multiple Threonine Residues—The role of post-translational mechanisms like phosphorylation in regulatory processes or the effect of phosphorylation on a given substrate in the physiology and/or cell division represent key events to our understanding of the signaling mechanisms through serine/threonine phosphorylation. This usually requires the identification of the phosphorylated sites, which often remains very challenging. To identify which of the 31 threonine residues of the C. glutamicum MurC corresponded to the phosphorylated site(s), a mass spectrometry approach was used. This technique has recently been proven to be a method of choice for characterizing post-translational modifications such as phosphorylation (18, 27, 37). LC-ESI/MS/MS was applied for the identification of phosphorylated peptides and for the localization of phosphorylation sites in MurC. Purified MurC was subjected to in vitro phosphorylation by PknA with non-radioactive ATP, prior to resolving on SDS-PAGE, and in-gel digestion with either trypsin or chymotrypsin. Phosphorylated amino acid residues were assigned by peptide fragmentation in MS/MS: y and b daughter ions containing one phosphothreonine were associated with a neutral loss of phosphoric acid (-H3PO4, i.e. -98 Da). As detailed in Table 3, analysis of tryptic and chymotryptic digests allowed the characterization of six phosphorylation sites in MurC corresponding to Thr51, Thr120, Thr133, Thr167, Thr362, and Thr365 (Figs. 1 and 4). These residues were found in the three structural domains of the MurC protein, two of them, namely Thr120 and Thr133, being located very close to the ATP-binding site of the protein (129GKT131). Thus, MS profiling coupled to in vitro kinase assays unambiguously demonstrated the phosphorylation of MurC by the PknA kinase. The fact that threonines but not serine residues were identified was consistent with our phosphoamino analysis (Fig. 2B).
TABLE 3.
Phosphorylation status of recombinant MurC as determined by mass spectrometry
| Phosphorylated tryptic and chymotryptic peptide sequence | Number of detected phosphate groups | Phosphorylated residue(s) |
|---|---|---|
| LC-ESI/MS/MS | ||
| [348-372] FNGAAITDDYAHHPTEVTAVLSAAR | 1 | Thr362 |
| [348-372] FNGAAITDDYAHHPTEVTAVLSAAR | 1 | Thr365 |
| [348-372] FNGAAITDDYAHHPTEVTAVLSAAR | 2 | Thr362 and Thr365 |
| [40-56] TVTGSDAKDSRTLLPLR | 1 | Thr51 |
| [48-56] DSRTLLPLR | 1 | Thr51 |
| [108-130] RSDLLGELLEGSTQVLIAGTHGK | 1 | Thr120 |
| [109-130] SDLLGELLEGSTQVLIAGTHGK | 1 | Thr120 |
| [159-184] AGTNAHHGTGEVFIAEADESDASLLR | 1 | Thr167 |
| [131-158] TSTTSMSVVAMQAAGMDPSFAIGGQLNK | 1 | Thr133 |
| [124-141] IAGTHGKTSTTSMSVVAM | 1 | Thr133 |
Definitive identification and localization of the six threonine residues identified by mass spectrometry was achieved by site-directed mutagenesis by introducing mutations that prevent their specific phosphorylation. Thus, all the six threonine residues were sequentially replaced by alanine, yielding the MurC_1T to MurC_6T mutants. These mutant proteins were expressed, purified as His-tagged proteins in E. coli BL21(DE3) Star harboring the different mutant constructs (Table 1), and used in an in vitro kinase assay. After TEV protease cleavage of the His tag, the recombinant MurC phosphorylation site mutants were incubated with [γ-33P]ATP and PknA. The proteins were separated by SDS-PAGE and analyzed by autoradiography. As shown in Fig. 4B (upper panel), equal amounts of MurC_WT or MurC mutants were used. Phosphorylation of the MurC_6T protein in which all six threonine residues were replaced by alanine appeared completely abolished, compared with phosphorylation of the intermediate mutants, as evidenced by the absence of a specific radioactive band (Fig. 4B, lower panel). Thus, these results unambiguously demonstrated that MurC without its six threonine phosphorylation sites has lost its ability to be phosphorylated by PknA, thus confirming the identification of all the sites of phosphorylation. An additional round of mass spectrometry analysis was also performed directly on the MurC_6T protein, which failed to identify any additional phosphate group that could eventually have arisen from a compensatory mechanism to the loss of the specific phosphorylation sites.
This type of analysis provided us the essential groundwork for mechanistic/functional studies of MurC regulation and demonstrated the efficiency of combining genetics and mass spectrometry analyses, with precise identification of phosphoacceptors, a prerequisite for a further understanding of the MurC mode of action. In particular, strains with defined mutations within the phosphorylation sites will be extremely helpful to establish the role of MurC phosphorylation-dependent regulation in corynebacterial growth and physiology.
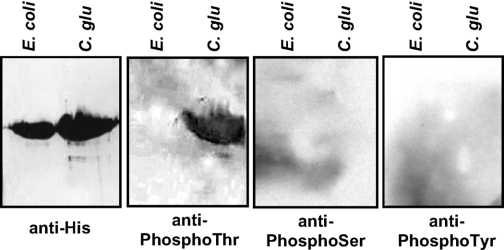
In Vivo Phosphorylation of MurC in C. glutamicum—To assess the relevance of in vitro phosphorylation, the in vivo phosphorylation of MurC was also investigated in C. glutamicum. Therefore, to determine whether phosphorylation of MurC occurs on threonine residues in vivo, Western blot analysis was performed using specific anti-phosphothreonine, anti-phosphoserine, anti-phosphotyrosine, or anti-His antibodies. To overproduce and purify phosphorylated MurC, an His tag was attached at the C terminus of the MurC protein under the control of the C. glutamicum divIVA promoter (15) in plasmid pEDiv (Table 1), and the recombinant protein was expressed in C. glutamicum. Cultures of E. coli_pTEVmurCor C. glutamicum_pEDivmurHis overexpressing the His-tagged MurC protein were collected and lysed, and the soluble MurC was then purified to homogeneity as previously described (21). It was found that MurC purified from the C. glutamicum strain was only phosphorylated on threonine residues (Fig. 5). This was consistent with our phosphoamino acid content analyses and the in vitro identification of the MurC phosphorylation sites when phosphorylated by PknA. It clearly establishes that the murein ligase MurC is highly phosphorylated in vivo on threonines. In contrast, the anti-phosphothreonine antibodies did not react with recombinant MurC protein purified from E. coli thus confirming that phosphorylation of the C. glutamicum MurC did not occur following heterologous expression in E. coli, which thus confirmed its specific phosphorylation in C. glutamicum (Fig. 5).
FIGURE 5.
In vivo phosphorylation of MurC in C. glutamicum. Recombinant MurC purified from either E. coli or C. glutamicum strains were analyzed by SDS-PAGE and detected by immunoblotting using antibodies against His tag, phosphothreonine, phosphoserine, or phosphotyrosine residues.
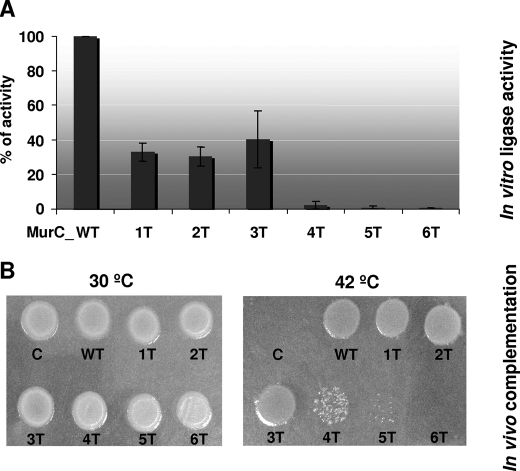
MurC Phosphorylation Sites Are Critical for in Vitro and in Vivo Activity—The importance of the phosphorylation sites for MurC ligase activity was investigated in vitro by site-directed mutagenesis. The six threonine residues were replaced by alanine, and the catalytic activity of the wild-type and mutant purified MurC proteins was assayed by following the incorporation of l-[14C]alanine into the PG nucleotide precursor, as described under “Experimental Procedures.” As shown in Fig. 6A, the activity of mutants MurC_1T, MurC_2T, and MurC_3T, corresponding to successive mutagenesis of Thr362, Thr365, and Thr51 residues, was reduced by ∼30-40% as compared with the wild-type level. Remarkably, the activity of the MurC_4T (T362A/T365A/T51A/T120A), MurC_5T (T362A/T365A/T51A/T120A/T167A), and MurC_6T (T362A/T365A/T51A/T120A/T167A/T133A) mutants appeared much more drastically altered and almost completely abolished in the six-threonine mutant (Fig. 6A). Moreover, to discern whether some threonines may be more important to murC activity than others individual mutagenesis was performed. The ligase activity of each individual mutant corresponding to MurC_T51A, MurC_T120A, MurC_T133A, MurC_T167A, MurC_T362A, and MurC_T365A, was of 72%, 29%, 44%, 79%, 26%, and 95% as compared with the wild-type level. Therefore, these results confirmed that Thr120, Thr133, and Thr362 represent critical residues for MurC ligase activity, as already observed with the MurC_1T to MurC_6T mutants proteins (Fig. 6A).
FIGURE 6.
Effect of MurC phosphorylation site mutagenesis on in vitro and in vivo activity. A, in vitro assays of the wild-type and phosphorylation site mutant (1T to 6T) MurC proteins. Purified MurC proteins were assayed as described under “Experimental Procedures.” Three independent experiments were performed, yielding similar results. The activity of the wild-type protein (here represented as 100%) was 2650 nmol/min/mg of protein, and those of the 1T, 2T, 3T, 4T, 5T, and 6T MurC mutants were 875, 800, 1050, 75, 20, and 10 nmol/min/mg of protein, respectively. B, in vivo functional complementation assays using an E. coli temperature-sensitive murC mutant strain. The pTrc99A plasmid vector and derivative plasmids expressing wild-type or mutated versions of the C. glutamicum MurC protein were transformed into the E. coli thermosensitive murC mutant strain H1119. Functional complementation was assayed by following the growth of the transformants at the permissive temperature of 30 °C (left panel), or at the non-permissive temperature of 42 °C (right panel).
These results confirmed that the overall activity of MurC relies on the presence of these threonine residues. In fact, one could imagine that replacing these critical residues present in all three characteristic domains of MurC could have a dramatic effect on the enzymatic activity of the ligase, and especially the mutagenesis of Thr133, a key residue closed to the GKT motif necessary for the binding of ATP.
The activity of the MurC proteins was also tested in vivo using a functional assay based on complementation of the E. coli temperature-sensitive murC mutant strain H1119. This strain, which grows normally at 30 °C but lyses when shifted at the non-permissive temperature of 42 °C, was transformed with the pTrc99A plasmid, or the pTrc99murC and pTrc99murC1T, 2T, 3T, 4T, 5T, and 6T derivatives (Table 1) expressing wild-type or mutated forms of MurC. As shown in Fig. 6B, all types of transformants grew normally at the permissive temperature of 30 °C, but only those expressing the wild-type protein or the 1T, 2T, and 3T mutated proteins restored growth of the E. coli mutant at the temperature of 42 °C. No complementation by the plasmids expressing the 4T, 5T, and 6T mutants was observed (Fig. 6B).
A perfect correlation was thus observed between the in vitro and in vivo data, i.e. between the catalytic activity of these proteins and their ability to complement the murC temperature-sensitive E. coli mutant. Combined together, the results obtained with the different phosphorylation site mutants stress the importance of these threonine residues for MurC activity. These data not only suggest that these residues are important for a fully expressed activity of the MurC protein, due probably to their localization in critical regions of the enzyme, but also indicate, if we put forward the analysis, that the introduction of a negative charge due to the phosphorylation of threonines may have an impact on the regulation of the MurC enzyme activity.
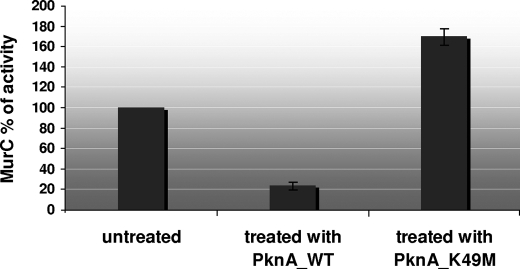
Phosphorylation Negatively Modulates MurC Ligase Activity—Therefore, we assessed whether phosphorylation by PknA could have a direct effect on the regulation of the ligase activity of MurC. The C. glutamicum MurC protein purified from E. coli was pre-treated with either the PknA_WT kinase or the PknA_K49M kinase, yielding the phosphorylated and non-phosphorylated isoforms of MurC, respectively, which were subsequently tested for activity (Fig. 7). The PknA_K49M kinase, which is unable to autophosphorylate (Fig. 3) and therefore transphosphorylate its substrate, represented here an appropriate control, because one could imagine that the kinase itself could be responsible for the effect on MurC activity. Interestingly, we found that the ligase activity of the phosphorylated isoform of MurC was severely inhibited, representing ∼30% of the wild-type activity, whereas no decrease of activity was observed following treatment by the PknA_K49M kinase (Fig. 7). An apparent increase of the MurC activity was observed in the latter case, suggesting that the presence of the mutant PknA protein, although inactive, stabilized in some way the MurC protein (protein-protein interactions) and reduced the slight loss of activity observed for MurC during the preincubation period. These results were consistent with the in vitro and in vivo analyses performed with the MurC mutants indicating that specific threonine residues were important in terms of enzymatic activity. Therefore, the PknA-mediated phosphorylation of MurC seems to represent a key mechanism for regulating/controlling its activity. Phosphorylation may rather modulate MurC activity on a fine-tuned level rather than a strict on/off mechanism. However, further work is needed to understand at a molecular level whether and how phosphorylation of MurC modifies the overall structure of this protein and negatively regulates the binding of l-alanine to UDP-MurNAc to generate the peptide moiety of the PG disaccharide peptide monomer unit.
FIGURE 7.
Comparative enzymatic activities of MurC phosphorylated versus non-phosphorylated. Purified wild-type MurC protein was phosphorylated either with PknA or PknA_K49M, and MurC ligase activity was assayed as described under “Experimental Procedures.” Two independent protein preparations were assayed in triplicate, yielding similar results.
We have previously shown that C. glutamicum PknA and PknB are key players in signal transduction pathways for the regulation of the cell shape and are both essential for sustaining corynebacterial growth (18). In this study, we extended our previous findings and demonstrated for the first time that PknA is able to specifically phosphorylate the essential murein ligase enzyme MurC, both in vitro and in vivo, and that post-translational modifications appear to be a way to regulate its ligase activity. Moreover, due to the fact that corynebacteria are extensively used for industrial production of amino acids such as glutamate, these results may open the way to manipulation of the cell wall chemical structure to increase amino acid secretion. Furthermore, these findings could be useful to extend our present understanding of the dcw (division-cell wall biosynthesis) cluster in Gram-positive bacteria toward designing of new antimicrobial drugs targeting the cell wall metabolism processes like the one involving the Mur enzymes. Moreover, if these inhibitors are capable of interfering with these regulatory processes through selective inhibition of PknA or MurC phosphorylation, it may lead to the design of novel classes of inhibitors. Indeed, the development of STPK inhibitors and the expertise in designing such inhibitors for eukaryotic STPKs represent an intense research area and could be exploited for the development of new drugs, especially toward the pathogenic corynebacteria Corynebacterium diphtheriae.
This work was supported in part by grants from the Région Rhône-Alpes (to M. C.); by the CNRS, the University of Lyon (France), and the National Research Agency (ANR-06-MIME-027-01 to V. M.); by the Junta de Castilla y León (Ref. LE040A07 to J. A. G.); by the Ministerio de Ciencia y Tecnología (Spain) (Grants BIO2008-00519 and BIO2005-02723 to J. A. G. and L. M. M.); and by the CNRS and Université Paris-Sud XI (Grant UMR 8619 to D. M. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PG, peptidoglycan; MurNAc, N-acetylmuramic acid; STPK, Ser/Thr protein kinase; GST, glutathione S-transferase; MS, mass spectrometry; MS/MS, tandem MS; LC, liquid chromatography; ESI, electrospray ionization; TEV, tobacco etch virus.
References
- 1.Vicente, M., Hodgson, J., Massidda, O., Tonjum, T., Henriques-Normark, B., and Ron, E. Z. (2006) FEMS Microbiol. Rev. 30 841-852 [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, M. D., Ross, H. O., Hillman, M. C., Meade, R. P., Kurilla, M. G., and Pompliano, D. L. (2002) Anal. Biochem. 306 17-22 [DOI] [PubMed] [Google Scholar]
- 3.Barreteau, H., Kovac, A., Boniface, A., Sova, M., Gobec, S., and Blanot, D. (2008) FEMS Microbiol. Rev. 32 168-207 [DOI] [PubMed] [Google Scholar]
- 4.van Heijenoort, J. (1996) in Escherichia Coli and Salmonella: Cellular and Molecular Biology (Press, A., ed) pp. 1025-1034, ASM Press, Washington, DC
- 5.van Heijenoort, J. (2001) Nat. Prod. Rep. 18 503-519 [DOI] [PubMed] [Google Scholar]
- 6.Mengin-Lecreulx, D., Texier, L., Rousseau, M., and van Heijenoort, J. (1991) J. Bacteriol. 173 4625-4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotnik, M., Anderluh, P. S., and Prezelj, A. (2007) Curr. Pharm. Des. 13 2283-2309 [DOI] [PubMed] [Google Scholar]
- 8.El Zoeiby, A., Sanschagrin, F., and Levesque, R. C. (2003) Mol. Microbiol. 47 1-12 [DOI] [PubMed] [Google Scholar]
- 9.Ehmann, D. E., Demeritt, J. E., Hull, K. G., and Fisher, S. L. (2004) Biochim. Biophys. Acta 1698 167-174 [DOI] [PubMed] [Google Scholar]
- 10.Zawadzke, L. E., Norcia, M., Desbonnet, C. R., Wang, H., Freeman-Cook, K., and Dougherty, T. J. (2008) Assay Drug Dev. Technol. 6 95-103 [DOI] [PubMed] [Google Scholar]
- 11.Strancar, K., Blanot, D., and Gobec, S. (2006) Bioorg. Med. Chem. Lett. 16 343-348 [DOI] [PubMed] [Google Scholar]
- 12.Strancar, K., Boniface, A., Blanot, D., and Gobec, S. (2007) Arch. Pharm. (Weinheim) 340 127-134 [DOI] [PubMed] [Google Scholar]
- 13.Hermann, T. (2003) J. Biotechnol. 104 155-172 [DOI] [PubMed] [Google Scholar]
- 14.Daniel, R. A., and Errington, J. (2003) Cell 113 767-776 [DOI] [PubMed] [Google Scholar]
- 15.Letek, M., Ordonez, E., Vaquera, J., Margolin, W., Flardh, K., Mateos, L. M., and Gil, J. A. (2008a) J. Bacteriol. 190 3283-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letek, M., Fiuza, M., Ordonez, E., Villadangos, A. F., Ramos, A., Mateos, L. M., and Gil, J. A. (2008b) Antonie Van Leeuwenhoek 94 99-109 [DOI] [PubMed] [Google Scholar]
- 17.Bendt, A. K., Burkovski, A., Schaffer, S., Bott, M., Farwick, M., and Hermann, T. (2003) Proteomics 3 1637-1646 [DOI] [PubMed] [Google Scholar]
- 18.Fiuza, M., Canova, M. J., Zanella-Cleon, I., Becchi, M., Cozzone, A. J., Mateos, L. M., Kremer, L., Gil, J. A., and Molle, V. (2008) J. Biol. Chem. 283 18099-18112 [DOI] [PubMed] [Google Scholar]
- 19.Thakur, M., and Chakraborti, P. K. (2008) Biochem. J. 415 27-33 [DOI] [PubMed] [Google Scholar]
- 20.Mateos, L. M., Schafer, A., Kalinowski, J., Martin, J. F., and Puhler, A. (1996) J. Bacteriol. 178 5768-5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molle, V., Brown, A. K., Besra, G. S., Cozzone, A. J., and Kremer, L. (2006) J. Biol. Chem. 281 30094-30103 [DOI] [PubMed] [Google Scholar]
- 22.Amann, E., Ochs, B., and Abel, K. J. (1988) Gene (Amst.) 69 301-315 [DOI] [PubMed] [Google Scholar]
- 23.Lugtenberg, E. J., and v Schijndel-van Dam, A. (1972) J. Bacteriol. 110 35-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liger, D., Masson, A., Blanot, D., van Heijenoort, J., and Parquet, C. (1995) Eur. J. Biochem. 230 80-87 [DOI] [PubMed] [Google Scholar]
- 25.Bouhss, A., Mengin-Lecreulx, D., Blanot, D., van Heijenoort, J., and Parquet, C. (1997) Biochemistry 36 11556-11563 [DOI] [PubMed] [Google Scholar]
- 26.Deva, T., Baker, E. N., Squire, C. J., and Smith, C. A. (2006) Acta Crystallogr D. Biol. Crystallogr. 62 1466-1474 [DOI] [PubMed] [Google Scholar]
- 27.Canova, M. J., Veyron-Churlet, R., Zanella-Cleon, I., Cohen-Gonsaud, M., Cozzone, A. J., Becchi, M., Kremer, L., and Molle, V. (2008) Proteomics 8 521-533 [DOI] [PubMed] [Google Scholar]
- 28.Molle, V., Kremer, L., Girard-Blanc, C., Besra, G. S., Cozzone, A. J., and Prost, J. F. (2003) Biochemistry 42 15300-15309 [DOI] [PubMed] [Google Scholar]
- 29.Peirs, P., De Wit, L., Braibant, M., Huygen, K., and Content, J. (1997) Eur. J. Biochem. 244 604-612 [DOI] [PubMed] [Google Scholar]
- 30.Huse, M., and Kuriyan, J. (2002) Cell 109 275-282 [DOI] [PubMed] [Google Scholar]
- 31.Johnson, L. N., Noble, M. E., and Owen, D. J. (1996) Cell 85 149-158 [DOI] [PubMed] [Google Scholar]
- 32.Dasgupta, A., Datta, P., Kundu, M., and Basu, J. (2006) Microbiology 152 493-504 [DOI] [PubMed] [Google Scholar]
- 33.Boitel, B., Ortiz-Lombardia, M., Duran, R., Pompeo, F., Cole, S. T., Cervenansky, C., and Alzari, P. M. (2003) Mol. Microbiol. 49 1493-1508 [DOI] [PubMed] [Google Scholar]
- 34.Hanks, S. K., Quinn, A. M., and Hunter, T. (1988) Science 241 42-52 [DOI] [PubMed] [Google Scholar]
- 35.Motley, S. T., and Lory, S. (1999) Infect. Immun. 67 5386-5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villarino, A., Duran, R., Wehenkel, A., Fernandez, P., England, P., Brodin, P., Cole, S. T., Zimny-Arndt, U., Jungblut, P. R., Cervenansky, C., and Alzari, P. M. (2005) J. Mol. Biol. 350 953-963 [DOI] [PubMed] [Google Scholar]
- 37.Molle, V., Zanella-Cleon, I., Robin, J. P., Mallejac, S., Cozzone, A. J., and Becchi, M. (2006) Proteomics 6 3754-3766 [DOI] [PubMed] [Google Scholar]
- 38.Schafer, A., Kalinowski, J., Simon, R., Seep-Feldhaus, A. H., and Puhler, A. (1990) J. Bacteriol. 172 1663-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santamaria, R. I., Martin, J. F., and Gil, J. A. (1987) Gene (Amst.) 56 199-208 [DOI] [PubMed] [Google Scholar]
- 40.Cohen-Gonsaud, M., Barthe, P., Pommier, F., Harris, R., Driscoll, P. C., Keep, N. H., and Roumestand, C. (2004) J. Biomol. NMR 30 373-374 [DOI] [PubMed] [Google Scholar]