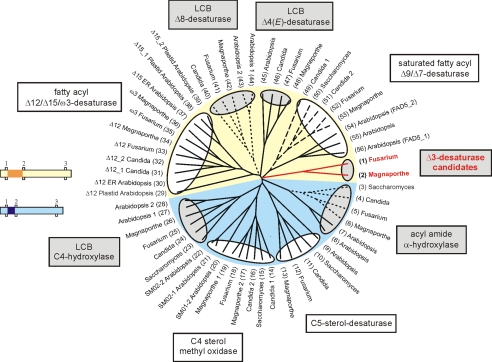
FIGURE 3.
Dendrogram showing desaturase-like sequences from F. graminearum, M. grisea, Candida albicans, S. cerevisiae, and A. thaliana. Sphingolipid-modifying enzymes are shaded in gray, and the Δ3(E)-desaturase candidates are marked in red. Cytochrome b5-fusion proteins are indicated by dashed lines (C-terminal fusion) and dotted lines (N-terminal fusion), respectively. The yellow and light blue background indicates groups of desaturase and hydroxylase sequences distinguished by a different spacing between the first and the second histidine box. The small pictures illustrate the distance between these histidine boxes, which is 25-34 amino acids in the sequences with yellow background and 7-18 amino acids in the sequences with light blue background. The spacing is highlighted in dark yellow and dark blue, respectively. GenBank protein accession numbers are: 1) XP_383758, 2) XP_368852, 3) NP_013999, 4) XP_717303, 5) XP_385340, 6) XP_361446, 7) NM_129030, 8) NM_11820, 9) NP_192948, 10) NP_013157, 11) XP_713612, 12) XP_382678, 13) XP_363987, 14) XP_722703, 15) NP_011574, 16) XP_713456, 17) XP_360827, 18) XP_390006, 19) XP_369331, 20) NP_567670, 21) NP_850133, 22) NP_563789, 23) NP_010583, 24) XP_715359, 25) XP_382698, 26) XP_369735, 27) AAF43928, 28) AAG52550, 29) NP_194824, 30) NP_850139, 31) XP_722258, 32) XP_714324, 33) XP_385960, 34) XP_365283, 35) XP_388066, 36) XP_362963, 37) NP_180559, 38) NP_187727, 39) NP_196177, 40) XP_719958, 41) XP_390021, 42) XP_361026, 43) NP_182144, 44) AAD00895, 45) NP_192402, 46) XP_722116, 47) XP_390550, 48) XP_001404401, 49) XP_714854, 50) NP_011460, 51) XP_717653, 52) XP_386360, 53) XP_363864, 54) NP_565721, 55) BAC43716, and 56) NP_172098. ER, endoplasmic reticulum; LCB, long-chain base.