Fig. 2.
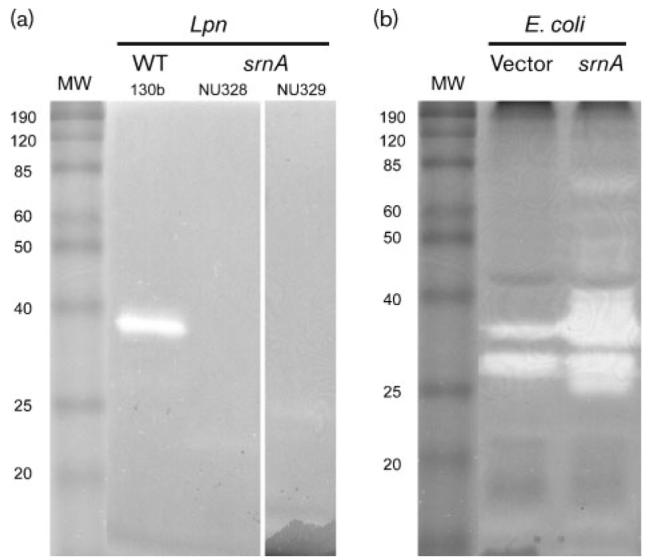
SrnA in L. pneumophila (Lpn) supernatants and recombinant E. coli lysates. (a) Neat supernatants obtained from wild-type (WT) 130b, and srnA mutants NU328 and NU239, were subjected to SDS-PAGE in the presence of RNA substrate, and then RNase was detected as a clear band following toluidine blue staining of the gel. MW, molecular mass markers (kDa) (BenchMark Protein Ladder; Invitrogen). The results depicted were obtained on two separate occasions using neat supernatants, and a further two times using 40× concentrated supernatants. (b) Lysates obtained from E. coli C41 (DE3) containing pET28α (vector) or pETsrnA that had srnA cloned into pET28α (srnA) were subjected to SDS-PAGE, and then RNase staining was done as described above. The two bands in the vector control lane probably represent endogenous E. coli RNase(s), whereas the band of approximately 25 kDa that is present in the sample from the srnA clone is probably a breakdown product of 38 kDa SrnA; results similar to each other were obtained on two separate occasions.
