Abstract
Background and Objective
The purpose of this study was to determine any difference between Porphyromonas gingivalis isolates from periodontally healthy sites as compared to those from diseased sites with respect to the ability to invade host cells.
Material and Methods
Subgingival plaque samples were obtained from periodontally healthy and diseased sites using paper points. P. gingivalis colonies were isolated and tested, using an antibiotic protection assay, for their ability to invade KB cells. P. gingivalis 381 and Escherichia coli MC1061 were used as controls.
Results
Mean values of 16.79 ± 0.86 × 103 colony-forming units/mL and 26.14 ± 2.11 × 103 colony-forming units/mL were observed in invasion assays for isolates from periodontally healthy and diseased sites, respectively. P. gingivalis present in diseased sites had significantly greater invasive abilities than strains isolated from healthy sites. No statistical difference was noted between male or female subjects concerning the degree of invasion; isolates from diseased sites from both genders had significantly greater invasion abilities than those from healthy sites. A significant correlation was found between the increased invasive capabilities of P. gingivalis isolates vs. an increased probing depth.
Conclusion
The increased invasion noted with P. gingivalis isolates from diseased sites vs. healthy sites, and the increased invasive capabilities with increasing probing depth, indicate that P. gingivalis isolates have a varying ability to invade host cells in the periodontal pocket.
Keywords: Porphyromonas gingivalis, invasion, periodontal disease, virulence
Periodontitis is a disease of the supporting structures of the teeth, causing loss of attachment to the alveolar bone with eventual exfoliation of teeth (1). Severe, generalized periodontitis affects up to 20% of the population, although mild-to-moderate periodontitis is observed in a majority of adults (2). Gram-negative bacteria play an important role in the pathogenesis of human periodontal diseases (3–5) but Porphyromonas gingivalis is one of the species most strongly implicated in periodontal disease (6,7).
Several putative virulence factors have been identified for P. gingivalis. These include extracellular proteolytic enzymes that can degrade host tissues and immune response mediators, toxic metabolites and adherence factors that promote colonization (8). For example, Porphyromonas strains elaborate proteases with the potential to degrade opsonizing immunoglobulin and complement proteins (9,10). P. gingivalis has the capacity to inhibit neutrophil migration and possesses a capsule with antiphagocytic properties (11,12). Epithelial cells act as the first barrier of defence against bacteria, and the invasion of these cells is an early step in the establishment of infection by a microorganism. The invasion of oral epithelial cells by pathogenic bacteria thus probably represents an important virulence factor in the progression of periodontal disease, as the epithelial cells provide protection from the host immune system. In vitro, P. gingivalis has been shown to attach to and invade several cell types, including gingival epithelial cells and KB cells (13–17). KB cells were once thought to be derived from an oral cancer, but in fact they were found to be derived from a glandular cancer of the cervix (18). Nevertheless, KB cells have been used extensively as a model to study the interaction between P. gingivalis and oral host cells (13–15,17,19–29). In KB cells, P. gingivalis isolates have been classified as having high, moderate or low invasive properties, or to be non-invasive (30). P. gingivalis invasion of epithelial cells is correlated with the activation of calcium-dependent host cell signaling systems and has been reported to require the release of calcium from intracellular storage and a subsequent increase in the concentration of cytosolic calcium (31).
The tooth surface and the periodontal pocket harbor approximately 1010 bacteria per gram net weight (32). The composition of the subgingival microflora varies considerably in periodontal health and in various forms of periodontal disease. The pathogenic nature of P. gingivalis is suggested by the recovery of these organisms in higher proportions from progressive periodontitis lesions than from quiescent periodontal sites, as well as by the elevated antibody levels found against P. gingivalis (serum and gingival crevicular fluid) in patients with periodontitis compared to normal controls. Studies have shown that P. gingivalis occurs with greater frequency and at higher levels in sites that appear to be disease active (6,33–38) and that certain periodontal health indicators in individuals are inversely correlated with the presence or with the levels of P. gingivalis (39–43). However, other studies have suggested that P. gingivalis is found more frequently in periodontally healthy sites than in sites of periodontal disease (44,45).
The number of sites required for an accurate microbiological assessment of periodontitis patients can be significantly decreased by the selection of deep pockets (46). Sites within subjects do not appear to be actively progressing at all times, so that the time of sampling may play as critical a role as the site to be sampled for understanding the pathogenesis of disease (47).
Griffen et al. identified 11 heteroduplex types of P. gingivalis, some of which were more strongly associated with disease than others (48). An association between P. gingivalis strains harboring the prtC+ fimA+ genotype and predominance in deep pockets or serious attachment loss was also recently suggested (49). This indicates that virulence in human periodontitis varies among strains of P. gingivalis, and that subgroups of highly virulent strains may be present at diseased sites. The fact that there are major differences in the degree of virulence of different isolates of P. gingivalis suggests that in some instances when suspected pathogens are found in periodontally healthy sites, the strains may be avirulent. An inability to distinguish virulent from avirulent clonal types has impeded the understanding of the pathogenesis of disease. The purpose of this study was to determine if fresh isolates of P. gingivalis from the human oral cavity have varying invasive abilities, and, if so, if this variation is related to their presence in healthy and diseased periodontal sites.
Material and methods
Sampling methodology
Study population
This study was approved by the Institutional Review Board of the University of Florida. Subjects for this study were recruited from the clinics of the College of Dentistry at the University of Florida. Potential subjects were screened, examined and selected for participation if they met criteria for sites of both periodontal health and disease in their oral cavity. For the purposes of this study, periodontally healthy sites included pocket depths and attachment levels that were < 5.0 mm and did not bleed upon probing. Periodontal disease sites were those sites with loss of attachment and pocket depths of ≥ 5.0 mm and which bled upon probing. Probing depth measurements were recorded by a single examiner to the nearest 0.5 mm using a Michigan ‘O’ periodontal probe with Williams markings. Exclusion criteria for the study were: (i) antibiotic therapy within the previous 6 mo; (ii) oral prophylaxis or periodontal surgery in the sites to be sampled within the last 6 mo; (iii) current pregnancy; and (iv) patients with a history of diabetes, blood dyscrasias, or rheumatic fever.
Bacterial sampling
Excess saliva was removed using a sterile gauze pad to minimize the collection of transient contaminating bacteria from an exogenous source. Any apparent supragingival plaque was removed from the surface of the tooth to be sampled using a Gracey curet. A sterile, fine endodontic paper point (Caulk-Dentsply, Milford, DE, USA) was placed, using cotton pliers, in the sulcus of each site for each tooth to be tested until resistance was felt and then left in place for 10 s. Paper points were then placed in a Corning plastic tube (Corning Inc., Corning, NY, USA) containing 1.0 mL of pre-reduced, anaerobically sterilized Ringers transport media and then transported to the laboratory for processing. After a brief vortex, a series of 10-fold serial dilutions of each sample was prepared. The suspended bacteria were streaked onto tryptic soy agar (Difco Laboratories, Detroit, MI, USA) supplemented with 5% sheep blood (Lampire Biological Laboratories, Pipersville, PA, USA), 0.5% yeast extract (Difco), hemin (5 μg/mL) and menadione (5 μg/mL). The plates were then placed at 37°C in a Coy anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI, USA) with an atmosphere of 5% CO2, 10% H2 and 85% N2.
Identification of P. gingivalis isolates
Colonies that were black pigmented after 5–7 d of incubation were identified as P. gingivalis as follows: (i) they did not show red fluorescence under long-wave ultraviolet light; (ii) they had pigmentation in the center of the colony; and (iii) they were trypsin positive when tested with N-cL-benzoyl-L-arginine-7-amido-4-methylcoumarin HCl. Asingle P. gingivalis colony was randomly selected from each sample and restreaked onto agar plates as described above. Colony polymerase chain reaction (PCR) was performed to confirm these colonies as P. gingivalis strains. Briefly, one or two colonies were resuspended in QuickExtract DNA extraction solution (Epicenter, Madison, WI, USA) and incubated at 65°C and then at 98°C for 4 min each. The lysate solution was centrifuged and the supernatant analyzed by nested PCR using the Hot Star Taq DNA polymerase, as described previously, with specific primers for the 16S rRNA gene of P. gingivalis (50).
Bacterial strains and growth conditions
Bacterial subculture
P. gingivalis strain 381, originally isolated by A. Tanner (The Forsyth Institute, Boston, MA, USA), was subcultured on tryptic soy agar (Difco) supplemented with 5% sheep blood (Lampire), 0.5% yeast extract (Difco), hemin (5 μg/mL), vitamin K1 (5 μg/mL) and gentamicin (50 μg/mL) (Sigma Chemical Co., St Louis, MO, USA). All P. gingivalis cultures were incubated in the anaerobic chamber. Escherichia coli strain MC1061 (kindly provided by A.S. Bleiweis, Gainesville, FL, USA) was subcultured on Luria–Bertani medium consisting of Bacto-Agar (15 g/L; Difco), Bacto-Tryptone (10 g/L; Difco), yeast extract (5 g/L) and sodium chloride (10 g/L; Fisher Scientific, Springfield, NJ, USA) at 37°C aerobically.
Cell culture
KB cells (ATCC CCL-17) were maintained in Eagle’s minimal essential medium (Mediatech, Herndon, VA, USA) supplemented with 10% (v/v) fetal bovine serum (Hyclone Laboratories, Inc., Logan, UT, USA), 200 mM L-glutamine (Sigma) and 100 μg/mL of penicillin–streptomycin (Sigma). KB cells were cultured in 75-cm2 flasks (Sarstedt, Newton, NC, USA) at 37°C in a humidified atmosphere of 5% CO2. Confluent mono-layers of KB cells were split by treatment with Hank’s balanced salt solution (Mediatech) and trypsin-versene (BioWhittaker, Walkersville, MD, USA).
Antibiotic protection assay
The antibiotic protection assay used in this study was performed as described previously for gingival epithelial cells (26,51). Approximately 105 KB cells per well were seeded in 24-well tissue culture plates (Sarstedt), washed three times with phosphate-buffered saline and then infected by the addition of a resuspended, overnight broth culture of 107 bacteria per mL of antibiotic-free medium at 37°C. After 90 min of incubation, the medium was removed from the infected cells and the cells were washed three times with phosphate-buffered saline. Medium containing 300 μg/mL of gentamicin (Sigma) and 200 μg/mL of metronidazole (Sigma) was then added to each well and the plates were incubated aerobically at 37°C for an additional 60 min. Control wells containing no cells were also included to establish that the antibiotic treatment was effective at killing the extracellular bacteria. Finally, the medium was removed, the cells were washed three times with phosphate-buffered saline and the cells were lysed by the addition of sterile distilled water and incubated for 20 min at 37°C under aerobic conditions. Dilutions of the cell lysates infected with bacteria were plated in triplicate on agar plates and incubated. P. gingivalis 381 and E. coli MC1061 were used as positive and negative controls, respectively for each invasion assay. The numbers of colony-forming units of invasive bacteria were then enumerated.
Statistical analysis
A simple linear regression model was used to compare invasion eficiency for periodontally healthy vs. periodontal diseased sites. The relationship between degree of invasion eficiency was compared with gender differences and probing depths using analysis of variance. Pearson product moment correlations were used to determine the relationship between invasion and probing depth. P values of < 0.05 were considered significant. All values were enumerated as mean ± standard error of the mean (52).
Results
Thirty-six subjects (16 male, 20 female) provided bacterial samples for potential P. gingivalis isolation, of which 30 subjects (14 male, 16 female) had culturable P. gingivalis at both healthy and diseased sites. The results for both periodontally healthy and diseased sites (including gender, tooth site sampled, probing depth and average colony-forming units per milliliter), which were compiled from three independent invasion assays, were analysed. When tested in KB cells, the invasion efficiencies of the strains isolated during this study were primarily in the moderate invasion efficiency range, with a few samples in the low invasion efficiency range, according to the criteria previously described by Dorn et al. (30). P. gingivalis 381 (the positive control) and E. coli MC1061 (the negative control) demonstrated high invasion efficiency or non-invasiveness, respectively.
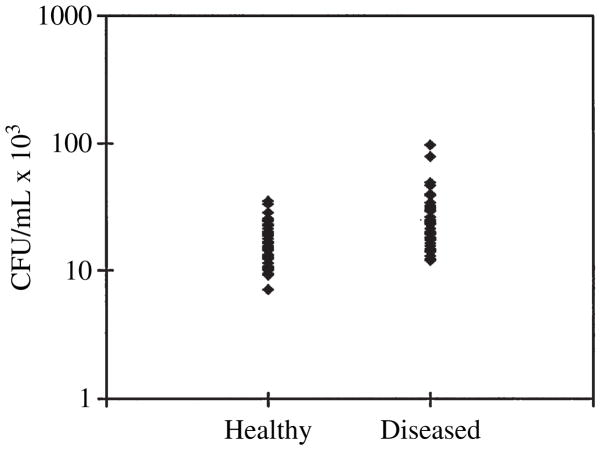
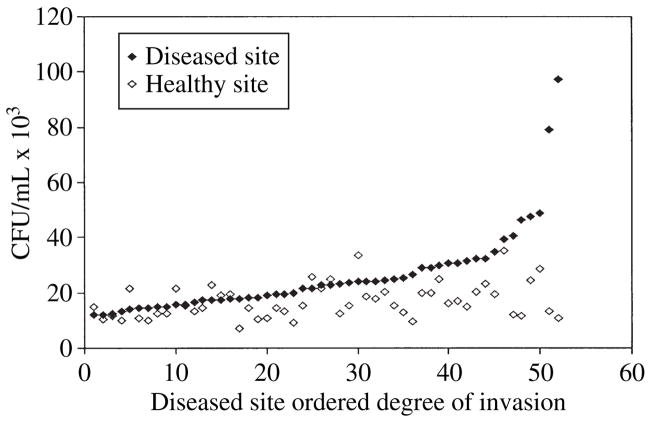
When a comparison was made between isolates from periodontally healthy and diseased sites, the mean degree of invasion for strains from periodontally healthy sites was 16.79 ± 0.86 × 103 colony-forming units/mL compared with 26.14 ±2.11 × 103 colony-forming units/mL for strains from periodontally diseased sites (Fig. 1). Thus, the level of invasion of strains from periodontally diseased sites was significantly higher (p < 0.0001) than the level of invasion of strains from the periodontally healthy sites. All strains with invasion frequencies in the low invasion range were isolated from healthy sites, while strains with the highest invasion level (at the high end of the moderate invasion efficiency category) were all isolated from periodontally diseased sites. While making a site-to-site comparison of degree of invasion within each patient, 82.7% of the diseased sites contained strains that exhibited a higher degree of invasion efficiency than the strains at healthy sites for each patient (Fig. 2). Thus, only 17.3% of healthy sites contained strains that exhibited slightly higher levels (average = 4000 (at the high end of the moderate invasion efficiency category) colony-forming units/mL higher) of invasion than their corresponding diseased site.
Fig. 1.
Invasion of KB cells by Porphyromonas gingivalis isolates from healthy and diseased periodontal sites. P. gingivalis isolates from subgingival plaque samples were tested, using an antibiotic protection assay, for their ability to invade KB cells. The mean level of invasion of strains isolated from periodontally healthy and diseased sites was 16.79 ± 0.86 × 103 colony-forming units/mL and 26.14 ± 2.11 × 103 colony-forming units/mL, respectively (p < 0.0001). CFU, colony-forming units.
Fig. 2.
Invasion of KB cells by Porphyromonas gingivalis from healthy and diseased sites. P. gingivalis isolates from healthy and diseased periodontal sites of each patient were compared for their ability to invade KB cells by using an antibiotic protection assay. The degree of invasion of P. gingivalis for each diseased site was recorded in increasing order along with the paired data from the corresponding healthy site. CFU, colony-forming units.
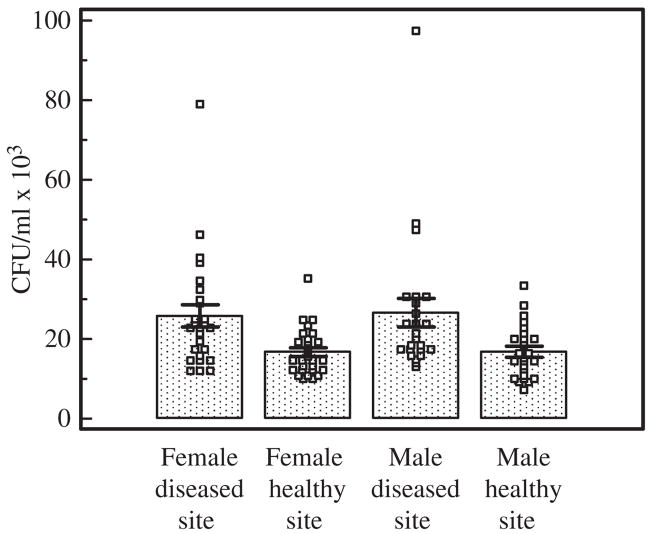
No statistical difference (p = 0.43) was noted between female (21.18 ± 1.60 colony-forming units/mL) and male (21.60 ± 2.01 colony-forming units/mL) subjects regarding degree of invasion. Both male and female subjects exhibited a strong difference in degree of invasion between healthy and diseased sites (p < 0.0001). The mean invasion efficiency of P. gingivalis for each probing depth was 16.76 ± 1.12 × 103 colony-forming units/mL for female healthy sites, 16.82 ± 1.33 × 103 colony-forming units/mL for male healthy sites, 25.53 ± 2.70 × 103 colony-forming units/mL for female diseased sites and 26.81 ± 3.48 × 103 colony-forming units/mL for male diseased sites (Fig. 3). These results indicate that the degree of P. gingivalis invasion efficiency is not gender dependent, but is dependent on the individual host/bacterial interactions.
Fig. 3.
Mean degree of invasion (± standard error of the mean) of KB cells by Porphyromonas gingivalis from healthy and diseased sites as grouped by gender. P. gingivalis isolates were tested, using an antibiotic assay, for their ability to invade KB cells and were then grouped by origin of the site (healthy vs. diseased and gender of the patient). Statistically significant differences were observed between both the male and female subjects vs. their healthy and diseased sites (p < 0.0001). CFU, colony-forming units.
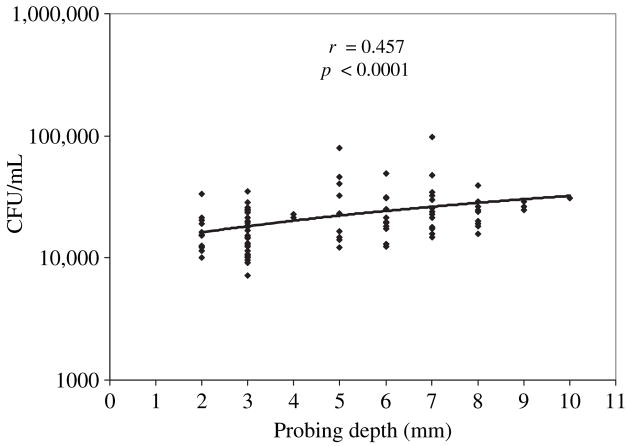
A significant correlation between probing depth and degree of invasion of P. gingivalis was observed. The coefficient of correlation (r) was 0.457 (95% confidence interval 0.290–0.597, p < 0.0001). (Fig. 4). The mean degree of invasion efficiency of P. gingivalis isolated from each probing depth was 16.69 ± 1.85 × 103 colony-forming units/mL for 2 mm, 16.55 ± 1.01 × 103 colony-forming units/mL for 3 mm, 22.08 ± 0.71 × 103 colony-forming units/mL for 4 mm, 30.16 ± 6.56 × 103 colony-forming units/mL for 5 mm, 23.36 ± 3.16 × 103 colony-forming units/mL for 6 mm, 28.75 ± 5.05 × 103 colony-forming units/mL for 7 mm, 20.94 ± 2.37 × 103 colony-forming units/mL for 8 mm, 26.66 ± 1.33 × 103 colony-forming units/mL for 9 mm and 30.70 × 103 colony-forming units/mL for 10 mm probing depth sites.
Fig. 4.
Degree of invasion of KB cells by Porphyromonas gingivalis vs. probing depth. P. gingivalis isolates were tested, using an antibiotic assay, for their ability to invade KB cells and were then grouped according to the probing depth recorded on the day of isolation. A significant correlation was found between the probing depth and the degree of invasion of P. gingivalis (p < 0.0001). CFU, colony-forming units.
Discussion
Animal test systems have shown that there are strain differences (clonality) in the virulence of P. gingivalis isolates (53–55). This is also observed with invasion (i.e. different laboratory strains exhibit varying invasion abilities). Dorn et al. (30) categorized P. gingivalis invasion efficiencies into four groups: high (×105), moderate (×104), low (×103) and noninvasive (≤ 102). Of 26 P. gingivalis strains tested by Dorn et al. (30), only P. gingivalis AJW4 was noninvasive, and P. gingivalis strain 381 possessed a 1000-fold greater ability than P. gingivalis AJW4 to invade KB cells. The invasion efficiencies obtained in this present study using fresh, clinical isolates were predominantly in the moderate range, with a few in the low range, and one strain from a disease site showed invasiveness close to the high efficiency level. Most periodontally diseased sites contained isolates that had greater invasion capabilities than did strains from the healthy sites, but some of the healthy sites contained strains with slightly greater invasion efficiencies than the corresponding diseased site. This increased invasion efficiency of strains from some healthy sites could be explained by the sites being in an ‘early disease stage’ at which the sites may be on the verge of a periodontal disease burst, but where sufficient attachment loss to classify the site as diseased, according to the criteria of the present study, has not yet occurred. Conversely, according to the burst hypothesis, the P. gingivalis strains present in the diseased sites may have, at the time of sampling, entered a quiescent phase of disease. In order to determine if this was the case, each diseased site in each patient would have to be evaluated longitudinally for disease progression related to the degree of P. gingivalis invasion efficiency. An additional and likely possibility for the difference in invasive efficiencies of strains between the healthy and diseased sites could be the presence of differing strains (clonal types) of P. gingivalis present at each site. Future studies will use genomic (such as arbitrary PCR) and phenotypic (such as serotyping, and measuring collagenolytic and proteolotic activities) analyses to determine if the isolates obtained from the healthy and diseased sites are in fact different strains. Griffen et al. (48) reported that the level of virulence among strains of P. gingivalis from human periodontitis varies, and that certain P. gingivalis subgroups are highly virulent. These virulent strains may be responsible for the attachment loss present at diseased sites in this study, whereas less virulent strains may be present in the same individual and may have colonized the healthy sites, explaining why these sites exhibit less attachment loss.
A significant correlation between probing depth and P. gingivalis degree of invasion was observed. As the pocket depth increases there are ecological changes in the pocket, which then result in changes within the biofilm. Investigations have previously shown that bacterial characteristics may be influenced by the surrounding biofilm. For example, Streptococcus cristatus, a gram-positive bacterium, is capable of modulating virulence expression of P. gingivalis through repression of the P. gingivalis fimbrial gene (fimA). Thus, P. gingivalis was unable to form biofilm microcolonies with S. cristatus (56). The biofilm interactions present at shallow probing depths may have an inhibitory effect on the invasive capabilities of P. gingivalis, whereas at deeper probing depths the bacteria present may assist the survival of more invasive P. gingivalis strains. In addition, host mechanisms may be present that select for strains which are more invasive at deeper probing depths and that may not be present in shallow pockets. Takemoto et al. found, using a rabbit disease model, that mono-inoculation with Tannerella forsythia (opinion on name change from Tannerella forsythensis pending; formerly Bacteroides forsythus), P. gingivalis or Fusobacterium nucleatum did not cause abscess formation. However, when T. forsythia was co-inoculated with either P. gingivalis or F. nucleatum, abscess formation occurred at 100% and 75% of the sites, respectively (57). Thus, biofilm interactions may trigger changes that modulate the survival/colonization of a particular P. gingivalis strain at deeper pocket depths, which may shift the predominant strains to those that are more invasive. Invariably, the host defense mechanisms may promote the emergence of P. gingivalis strains with increased invasion abilities and inevitably greater tissue destruction.
Adherence and invasion are active processes in which microorganisms may use host proteins and enzymes to gain entry into the cell, thus stimulating their own uptake. P. gingivalis has developed invasion strategies and mechanisms similar to those of other pathogens for both epithelial and endothelial cells. A common strategy among invasive bacteria is to trigger the host cell to undergo cytoskeletal rearrangements mediated by actin polymerization (58). P. gingivalis invasion has been shown to be inhibited by thapsigargin and 1,2-bis(2-aminophenoxy) ethane-N,N, N1,N1-tetraacetic acid (31), as well as by cytochalasin D, nocodazole, staurosporine, protease inhibitors and sodium azide, thus indicating that cytoskeletal rearrangements, protein phosphorylation, energy metabolism and P. gingivalis proteases are essential for invasion (51,59).
It seems logical to expect greater invasion efficiency of P. gingivalis in sites with evidence of periodontal disease, as this would support evidence of the ability of P. gingivalis to interact with the host tissues and thus cause disease. Countering the increased invasive abilities of P. gingivalis present in deeper periodontal pockets may require multiple subgingival environment disruptions via scaling and root planing, or may require the adjunctive use of subgingival antimicrobial or antibiotic modalities. In addition, monitoring of the subgingival flora for bacterial strains with greater invasive abilities may be required in order to predict the possibility of future disease progression and which methods of treatment would be best utilized to control the disease process.
In summary, periodontal sampling of P. gingivalis at healthy and diseased sites showed that P. gingivalis strains isolated from diseased sites possess greater invasion efficiencies than strains from healthy sites. This increased ability to invade cells may promote disease progression. Invasion efficiency differences were not found to be related to gender, but both male and female patients exhibited significantly greater P. gingivalis invasion at diseased sites. Also, greater invasion efficiency was noted for increasing probing depth.
Acknowledgments
We would like to thank Joan Whitlock for technical assistance. This study was supported by the National Institute of Dental and Craniofacial Research Grants DE 07496 and DE 013545.
References
- 1.Brown LJ, Oliver RC, Loe H. Periodontal diseases in the U.S. in 1981: prevalence, severity, extent, and role in tooth mortality. J Periodontol. 1989;60:363–370. doi: 10.1902/jop.1989.60.7.363. [DOI] [PubMed] [Google Scholar]
- 2.Burt B. Position paper: epidemiology of periodontal diseases. J Periodontol. 2005;76:1406–1419. doi: 10.1902/jop.2005.76.8.1406. [DOI] [PubMed] [Google Scholar]
- 3.Socransky SS. Microbiology of periodontal disease-present status and future considerations. J Periodontol. 1977;48:497–504. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- 4.Slots J. Subgingival microflora and periodontal disease. J Clin Periodontol. 1979;6:351–382. doi: 10.1111/j.1600-051x.1979.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 5.Haake SK, Meyer DH, Fives-Taylor PM, Schenkein HA. Periodontal diseases. Washington: ASM Press; 2006. [Google Scholar]
- 6.Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 8.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74:111–118. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- 10.O’Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003;4:409–426. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 11.Madianos PN, Papapanou PN, Sandros J. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect Immun. 1997;65:3983–3990. doi: 10.1128/iai.65.10.3983-3990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect Immun. 2006;74:5352–5361. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan MJ, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamont RJ, Oda D, Persson RE, Persson GR. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol Immunol. 1992;7:364–367. doi: 10.1111/j.1399-302x.1992.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 15.Lamont RJ, Yilmaz O. In or out: the invasiveness of oral bacteria. Periodontol 2000. 2002;30:61–69. doi: 10.1034/j.1600-0757.2002.03006.x. [DOI] [PubMed] [Google Scholar]
- 16.Sandros J, Papapanou P, Dahlen G. Porphyromonas gingivalis invades oral epithelial cells in vitro. J Periodontal Res. 1993;28:219–226. doi: 10.1111/j.1600-0765.1993.tb01072.x. [DOI] [PubMed] [Google Scholar]
- 17.Sandros J, Papapanou PN, Nannmark U, Dahlen G. Porphyromonas gingivalis invades human pocket epithelium in vitro. J Periodontal Res. 1994;29:62–69. doi: 10.1111/j.1600-0765.1994.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 18.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 19.Houalet-Jeanne S, Pellen-Mussi P, Tricot-Doleux S, Apiou J, Bonnaure-Mallet M. Assessment of internalization and viability of Porphyromonas gingivalis in KB epithelial cells by confocal microscopy. Infect Immun. 2001;69:7146–7151. doi: 10.1128/IAI.69.11.7146-7151.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umemoto T, Hamada N. Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis. J Periodontol. 2003;74:119–122. doi: 10.1902/jop.2003.74.1.119. [DOI] [PubMed] [Google Scholar]
- 21.Sojar HT, Genco RJ. Identification of glyceraldehyde-3-phosphate dehydrogenase of epithelial cells as a second molecule that binds to Porphyromonas gingivalis fimbriae. FEMS Immunol Med Microbiol. 2005;45:25–30. doi: 10.1016/j.femsim.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Tamai R, Asai Y, Ogawa T. Requirement for intercellular adhesion molecule 1 and caveolae in invasion of human oral epithelial cells by Porphyromonas gingivalis. Infect Immun. 2005;73:6290–6298. doi: 10.1128/IAI.73.10.6290-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eick S, Reissmann A, Rodel J, Schmidt KH, Pfister W. Porphyromonas gingivalis survives within KB cells and modulates inflammatory response. Oral Microbiol Immunol. 2006;21:231–237. doi: 10.1111/j.1399-302X.2006.00282.x. [DOI] [PubMed] [Google Scholar]
- 24.Pathirana RD, O’Brien-Simpson NM, Visvanathan K, Hamilton JA, Reynolds EC. Flow cytometric analysis of adherence of Porphyromonas gingivalis to oral epithelial cells. Infect Immun. 2007;75:2484–2492. doi: 10.1128/IAI.02004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umeda JE, Missailidis C, Longo PL, Anzai D, Wikstrom M, Mayer MP. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol. 2006;21:415–419. doi: 10.1111/j.1399-302X.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 26.Yuan L, Rodrigues PH, Bélanger M, Dunn W, Jr, Progulske-Fox A. The Porphyromonas gingivalis clpB gene is involved in cellular invasion in vitro and virulence in vivo. FEMS Immunol Med Microbiol. 2007;51:388–398. doi: 10.1111/j.1574-695X.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 27.Sandros J, Papapanou P, Dahlen G. Porphyromonas gingivalis invades oral epithelial cells in vitro [published erratum appears in J Periodontal Res 1993 Sep;28(5):386] J Periodontal Res. 1993;28:219–226. doi: 10.1111/j.1600-0765.1993.tb01072.x. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Lee SW, Hillman JD, Progulske-Fox A. Identification and testing of Porphyromonas gingivalis virulence genes with a pPGIVET system. Infect Immun. 2002;70:928–937. doi: 10.1128/IAI.70.2.928-937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eick S, Rodel J, Einax JW, Pfister W. Interaction of Porphyromonas gingivalis with KB cells: comparison of different clinical isolates. Oral Microbiol Immunol. 2002;17:201–208. doi: 10.1034/j.1399-302x.2002.170401.x. [DOI] [PubMed] [Google Scholar]
- 30.Dorn BR, Burks JN, Seifert KN, Progulske-Fox A. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol Let. 2000;187:139–144. doi: 10.1111/j.1574-6968.2000.tb09150.x. [DOI] [PubMed] [Google Scholar]
- 31.Izutsu KT, Belton CM, Chan A, et al. Involvement of calcium in interactions between gingival epithelial cells and Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;144:145–150. doi: 10.1111/j.1574-6968.1996.tb08521.x. [DOI] [PubMed] [Google Scholar]
- 32.Slots J, Listgarten MA. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988;15:85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 33.Socransky SS, Haffajee AD, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 34.Melvin WL, Assad DA, Miller GA, Gher ME, Simonson L, York AK. Comparison of DNA probe and ELISA microbial analysis methods and their association with adult periodontitis. J Periodontol. 1994;65:576–582. doi: 10.1902/jop.1994.65.6.576. [DOI] [PubMed] [Google Scholar]
- 35.Preus HR, Anerud A, Boysen H, Dunford RG, Zambon JJ, Loe H. The natural history of periodontal disease. The correlation of selected microbiological parameters with disease severity in Sri Lankan tea workers. J Clin Periodontol. 1995;22:674–678. doi: 10.1111/j.1600-051x.1995.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 36.Wolff LF, Aeppli DM, Pihlstrom B, et al. Natural distribution of 5 bacteria associated with periodontal disease. J Clin Periodontol. 1993;20:699–706. doi: 10.1111/j.1600-051x.1993.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J Periodontol. 2001;72:1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 38.Chen LL, Wu YM, Yan J, Sun WL, Sun YZ, Ojcius D. Association between coinfection of Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and Treponema denticola and periodontal tissue destruction in chronic periodontitis. Chin Med J (Engl) 2005;118:915–921. [PubMed] [Google Scholar]
- 39.Grossi SG, Zambon JJ, Ho AW, et al. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 40.Haffajee AD, Socransky SS, Smith C, Dibart S. Relation of baseline microbial parameters to future periodontal attachment loss. J Clin Periodontol. 1991;18:744–750. doi: 10.1111/j.1600-051x.1991.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 41.Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–739. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 42.Alpagot T, Wolff LF, Smith QT, Tran SD. Risk indicators for periodontal disease in a racially diverse urban population. J Clin Periodontol. 1996;23:982–988. doi: 10.1111/j.1600-051x.1996.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 43.Teanpaisan R, Douglas CW, Walsh TF. Characterisation of black-pigmented anaerobes isolated from diseased and healthy periodontal sites. J Periodontal Res. 1995;30:245–251. doi: 10.1111/j.1600-0765.1995.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 44.Wilson M, Lopatin D, Osborne G, Kieser JB. Prevalence of Treponema denticola and Porphyromonas gingivalis in plaque from periodontally-healthy and periodontallydiseased sites. J Med Microbiol. 1993;38:406–410. doi: 10.1099/00222615-38-6-406. [DOI] [PubMed] [Google Scholar]
- 45.Gornitsky M, Clark DC, Siboo R, et al. Clinical documentation and occurrence of putative periodontopathic bacteria in human immunodeficiency virus-associated periodontal disease. J Periodontol. 1991;62:576–585. doi: 10.1902/jop.1991.62.9.576. [DOI] [PubMed] [Google Scholar]
- 46.Gunsolley JC, Chinchilli VN, Savitt ED, et al. Analysis of site specific periodontal bacteria sampling schemes. J Periodontol. 1992;63:507–514. doi: 10.1902/jop.1992.63.6.507. [DOI] [PubMed] [Google Scholar]
- 47.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 48.Griffen AL, Lyons SR, Becker MR, Moeschberger ML, Leys EJ. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol. 1999;37:4028–4033. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu YM, Yan J, Chen LL, Gu ZY. Association between infection of different strains of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in subgingival plaque and clinical parameters in chronic periodontitis. J Zhejiang Univ Sci B. 2007;8:121–131. doi: 10.1631/jzus.2007.B0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boggess KA, Madianos PN, Preisser JS, Moise KJ, Jr, Offenbacher S. Chronic maternal and fetal Porphyromonas gingivalis exposure during pregnancy in rabbits. Am J Obstet Gynecol. 2005;192:554–557. doi: 10.1016/j.ajog.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montgomery D. Design and Analysis of Experiments. 2. New York: John Wiley & Sons Inc; 1984. [Google Scholar]
- 53.Grenier D, Mayrand D. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol. 1987;25:738–740. doi: 10.1128/jcm.25.4.738-740.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Steenbergen TJM, Delamarre FGA, Namavar F, de Graaf J. Differences in virulence within the species Bacteroides gingivalis. Antonie Van Leeuwenhoek. 1987;53:233–244. doi: 10.1007/BF00393930. [DOI] [PubMed] [Google Scholar]
- 55.Neiders ME, Chen PB, Suido H, et al. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodont Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 56.Xie H, Cook GS, Costerton JW, Bruce G, Rose TM, Lamont RJ. Intergeneric communication in dental plaque biofilms. J Bacteriol. 2000;182:7067–7069. doi: 10.1128/jb.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takemoto T, Kurihara H, Dahlen G. Characterization of Bacteroides forsythus isolates. J Clin Microbiol. 1997;35:1378–1381. doi: 10.1128/jcm.35.6.1378-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenshine I, Finlay BB. Exploitation of host signal transduction pathways and cytoskeletal functions by invasive bacteria. Bioessays. 1993;15:17–24. doi: 10.1002/bies.950150104. [DOI] [PubMed] [Google Scholar]
- 59.Deshpande RG, Khan MB, Genco CA. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun. 1998;66:5337–5343. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]