Summary
The nitric oxide (NO) cytotoxicity has been well documented in bacteria and mammalian cells. However, the underlying mechanism is still not fully understood. Here we report that transient NO exposure effectively inhibits cell growth of Escherichia coli in minimal medium under anaerobic growth conditions, and that cell growth is restored when the NO-exposed cells are either supplemented with the branched-chain amino acids (BCAA) anaerobically or returned to aerobic growth conditions. The enzyme activity measurements show that dihydroxyacid dehydratase (IlvD), an iron-sulfur enzyme essential for the BCAA biosynthesis, is completely inactivated in cells by NO with the concomitant formation of the IlvD-bound dinitrosyl iron complex (DNIC). Fractionation of the cell extracts prepared from the NO-exposed cells reveals that a large number of different protein-bound DNICs are formed by NO. While the IlvD-bound DNIC and other protein-bound DNICs are stable in cells under anaerobic growth conditions, they are efficiently repaired under aerobic growth conditions even without new protein synthesis. Additional studies indicate that L-cysteine may have an important role in repairing the NO-modified iron-sulfur proteins in aerobically growing E. coli cells. The results suggest that cellular deficiency to repair the NO-modified iron-sulfur proteins may directly contribute to the NO-induced bacteriostasis under anaerobic conditions.
Keywords: nitric oxide, bacteriostasis, iron-sulfur proteins, dinitrosyl iron complex
Introduction
As a neutral free radical, nitric oxide (NO) can readily penetrate cell membranes and act on a number of cellular components. At low concentrations (nM), NO is a signaling molecule for intercellular communications in neuronal and cardiovascular tissues (Ignarro, 1999). At high concentrations (μM), NO becomes a powerful weapon to kill pathogenic bacteria and tumor cells (Gobert et al., 2001; Krieglstein et al., 2001; MacMicking et al., 1997). In response to NO stress, bacteria have developed a number of defense mechanisms (Spiro, 2007). In Escherichia coli, there are at least three enzymes that can directly metabolize NO: a flavorubredoxin/flavodiiron NO reductase capable of reducing NO to nitrous oxide in the absence of oxygen (D'Autreaux et al., 2005; Gardner et al., 2002; Gomes et al., 2002), a flavohemoglobin NO dioxygenase that catalyzes oxidation of NO to nitrate in the presence of O2 (Bodenmiller and Spiro, 2006; Gardner and Gardner, 2002; Poole and Hughes, 2000), and a pentaheme nitrite reductase that has a NO reductase activity (Poock et al., 2002). In addition, a diiron protein encoded by gene ytfE has been shown to be critical for the cellular defense against NO (Justino et al., 2007). Deletion of any of these genes significantly increases the sensitivity of E. coli cells to NO under aerobic or anaerobic conditions. However, all these genes together are still not sufficient to protect E. coli from the NO cytotoxicity (Hyduke et al., 2007; Justino et al., 2005; Mukhopadhyay et al., 2004; Pullan et al., 2007).
Increasing evidence has suggested that proteins containing iron-sulfur clusters or mononuclear iron centers are susceptible to NO (Spiro, 2007). So far, over 200 unique iron-sulfur proteins have been identified in diverse physiological processes such as energy conversion, sugar metabolism, heme and biotin biosynthesis, amino acids biosynthesis, RNA modification, DNA synthesis and repair, and the regulation of gene expression (Johnson et al., 2005; Lill and Muhlenhoff, 2006). In vitro studies showed that purified iron-sulfur proteins can be readily modified by NO forming the protein-bound dinitrosyl iron complexes (DNICs) which have a unique electron paramagnetic resonance (EPR) signal at g = 2.04 (Cruz-Ramos et al., 2002; Ding and Demple, 2000; Drapier, 1997; Foster and Cowan, 1999; Kennedy et al., 1997; Rogers et al., 2003). The in vivo studies also indicated that iron-sulfur proteins such as the aconitase [4Fe-4S] clusters (Gardner et al., 1997), the ferredoxin [2Fe-2S] clusters (Rogers and Ding, 2001), the endonuclease III [4Fe-4S] clusters (Rogers et al., 2003), the redox transcription factor SoxR [2Fe-2S] clusters (Ding and Demple, 2000), the anaerobic growth factor FNR [4Fe-4S] clusters (Cruz-Ramos et al., 2002), the dihydroxyacid dehydratase (IlvD) [4Fe-4S] clusters (Hyduke et al., 2007), and a number of other dehydratase [4Fe-4S] clusters (Woodmansee and Imlay, 2003) are highly sensitive to NO. Additional evidence came from the micro-array gene profiling experiments which revealed that NO exposure drastically elevates the expression of the genes involved in the assembly and/or repair of iron-sulfur clusters in E. coli cells (Hyduke et al., 2007; Justino et al., 2005; Mukhopadhyay et al., 2004; Pullan et al., 2007). Collectively, these results suggested that iron-sulfur proteins may represent one of the primary targets of the NO cytotoxicity (Spiro, 2007). Nevertheless, the physiological relevance of the NO-mediated modifications of iron-sulfur proteins and the NO cytotoxicity has still not been fully addressed.
Here, we have explored the NO-induced bacteriostasis and modification of iron-sulfur proteins in E. coli. The results show that transient NO exposure effectively inhibits cell growth of E. coli in minimal medium under anaerobic growth conditions. However, cell growth is restored when the NO-exposed E. coli cells are either supplemented with the branched-chain amino acids (BCAA) anaerobically or returned to aerobic growth conditions. Consistent with the studies by Hyduke et al. (Hyduke et al., 2007), we find that dihydroxyacid dehydratase (IlvD), an iron-sulfur enzyme essential for the BCAA biosynthesis in E. coli (Flint et al., 1993), is a primary target of the NO cytotoxicity. The enzyme activity measurements in the cell extracts show that IlvD is completely inactivated by NO with the concomitant formation of the IlvD-bound DNIC in E. coli cells. Furthermore, we discover that the NO-inactivated IlvD remains inactive in E. coli cells under anaerobic growth conditions but is efficiently repaired under aerobic growth conditions even without new protein synthesis. We propose that intracellular small molecules may directly contribute to the robust repair activity for the NO-modified iron-sulfur proteins in E. coli under aerobic growth conditions. Additional studies reveal that L-cysteine may have an important role in repairing the NO-modified iron-sulfur proteins in aerobically growing E. coli cells. The results led us to suggest that NO may become a potent bacteriostat under anaerobic conditions under which bacterial cells fail to repair the NO-modified iron-sulfur proteins.
Results
NO-induced bacteriostasis of E. coli under anaerobic growth conditions
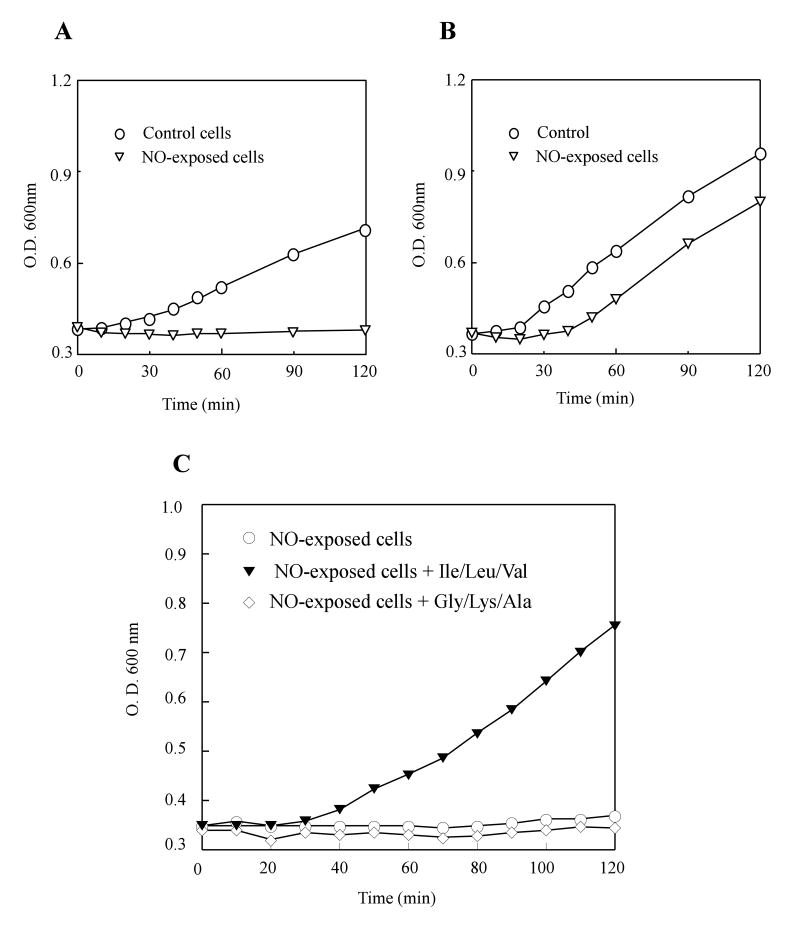
To explore the NO cytotoxicity in bacteria, we adapted the Silastic tubing NO delivery system (Tamir et al., 1993). The system allowed us to modulate a reproducible NO releasing by changing the length of the Silastic tubing immersed in the cell culture. Throughout the study, a releasing rate of about 100 nM NO per second was used to emulate the pathophysiological NO productions as reported by others (Gobert et al., 2001; Krieglstein et al., 2001). Typically, exponentially growing E. coli cells were purged with pure argon gas before being subjected to the NO exposure. After NO exposure for 10 min, E. coli cells were re-purged with pure argon gas to remove residual NO. The NO-exposed E. coli cells were then divided into two flasks, one to aerobic growth conditions and the other to anaerobic growth conditions. Figure 1A shows that the NO exposure effectively inhibited cell growth of E. coli in minimal medium under anaerobic growth conditions. On the other hand, when the NO-exposed E. coli cells were returned to aerobic growth conditions (Figure 1B), cell growth was restored with only a short delay as reported previously by Hyduke et al. (Hyduke et al., 2007).
Figure 1. NO-induced bacteriostasis of E. coli.
Exponentially growing E. coli cells were exposed to pure NO gas at a rate of ∼ 100 nM per second for 10 min anaerobically, followed by purge with pure argon gas to remove residual NO in cell culture. The NO-exposed E. coli cells were returned to either A) anaerobic or B) aerobic growth conditions in minimal medium containing 0.2% glucose. Circles: untreated E. coli cells. Triangles: the NO-exposed E. coli cells. C), the NO-exposed E. coli cells were returned to anaerobic growth conditions in minimal medium containing 0.2% glucose supplemented with three branched-chain amino acids (leucine, isoleucine, valine) (filled triangles), three non-branched-chain amino acids (glycine, alanine and lysine) (open diamonds) or an equal volume of de-gassed water (open circles) anaerobically. The final concentration for each amino acid in the minimal medium was 100 μg/ml. The cell growth was measured as described in the Experimental Procedures. The data are the representatives of three independent experiments.
The recovery of the NO-exposed E. coli cells under aerobic growth condition (Figure 1B) suggests that the NO exposure may have limited the production of some intermediates essential for cell growth under anaerobic growth conditions. Hyduke et al. (Hyduke et al., 2007) has suggested that the NO exposure may create a transient deficiency of the branched-chain amino acids (BCAA) in E. coli cells under aerobic growth conditions, thus contributing to a short delay of cell growth. To test whether the NO exposure results in a prolonged BCAA auxotrophy of E. coli under anaerobic growth conditions, we supplemented the minimal medium with various components and found that addition of BCAA (leucine, isoleucine and valine) largely alleviated the NO-induced inhibition of cell growth under anaerobic growth conditions (Figure 1C). In control, addition of the non-branched-chain amino acids (glycine, lysine and alanine) failed to restore cell growth of the NO-exposed E. coli cells under anaerobic growth conditions (Figure 1C). Thus, the results indicated that the NO exposure may have created the BCAA auxotrophy of E. coli under anaerobic growth conditions.
Dihydroxyacid dehydratase (IlvD) in E. coli cells is inactivated by NO
The BCAA auxotrophy is a hallmark of oxidative damage to iron-sulfur clusters in E. coli (Jang and Imlay, 2007) and in Saccharomyces cerevisiae (Wallace et al., 2004). The BCAA biosynthesis pathway in E. coli contains two iron-sulfur enzymes: dihydroxyacid dehydratase (IlvD) (Flint et al., 1993) and isopropylmalate isomerase (LeuCD) (Jang and Imlay, 2007). IlvD catalyzes the conversion from 2,3-dihydroxy-isovalerate to 2-keto-isovalerate (Flint et al., 1993), while LeuCD catalyzes the conversion from 2-isopropylmalate to 3-isopropylmalate (Jang and Imlay, 2007). Both IlvD and LeuCD require an intact [4Fe-4S] cluster to act as a Lewis acid for the catalytic function. Because LeuCD is a two-subunit enzyme that dissociates during purification (Jang and Imlay, 2007), we chose IlvD for further investigation.
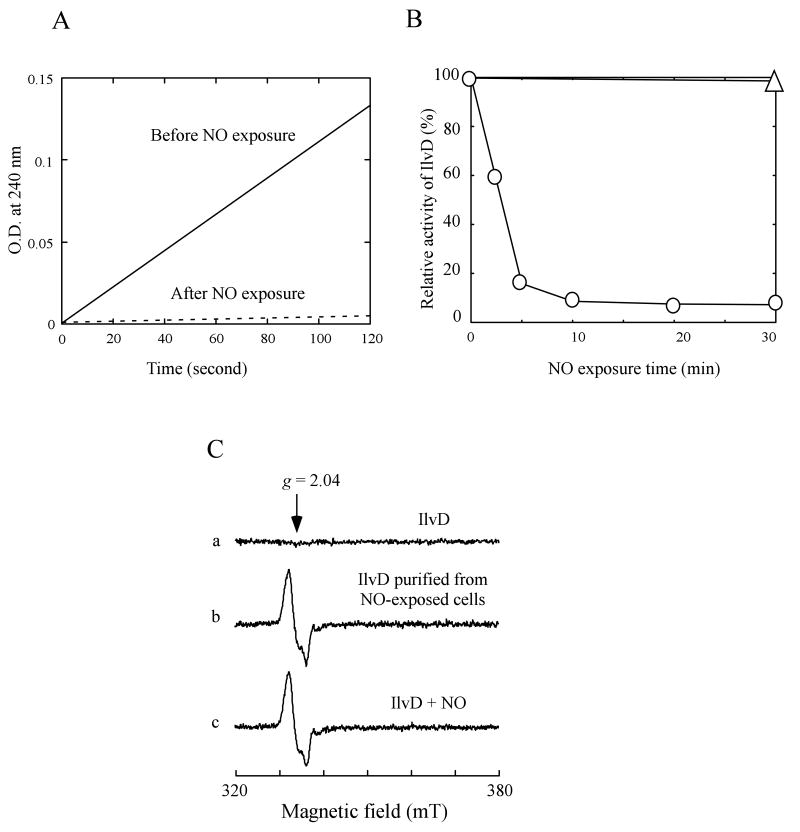
To facilitate the rapid enzyme activity assay in the cell extracts, we expressed recombinant IlvD to about 2% of total intracellular proteins in E. coli cells. Figure 2A shows that when E. coli cells containing recombinant IlvD were subjected to the NO exposure at a rate of 100 nM NO per second anaerobically, the enzyme activity of IlvD in cells was quickly inactivated, with over 80% of the IlvD activity was eliminated after 5 min NO exposure (Figure 2B). It is worth mentioning that the enzyme activity of IlvD in E. coli cells was completely inactivated after 10 min NO exposure using the Silastic tubing NO delivery system (Figure 2A), whereas at least 50% of the IlvD activity still remained when E. coli cells were exposed to the NO-releasing reagent DeaNO (8 μM) (Hyduke et al., 2007), indicating that the Silastic tubing NO delivery system may be more effective than the NO bolus exposure in treating bacterial cells.
Figure 2. Inactivation of dihydroxyacid dehydratase (IlvD) in E. coli cells by NO.
A), the enzyme activity of recombinant IlvD in E. coli cells before and after 10 min NO exposure. The cell extracts were prepared from the E. coli cells (concentrated to O.D. at 600 nm of 3.0) before and after the NO exposure. The reaction product (keto acids) catalyzed by IlvD was monitored at 240 nm as described in the Experimental Procedures. The enzyme activity of IlvD in untreated E. coli cells (100%) was ∼11.3 μmol ketos formed per mg total cellular protein per min. B), kinetics of the NO-mediated inactivation of IlvD in E. coli cells. Aliquots of the cell culture were taken at indicated time points during the NO exposure. The cell extracts were prepared from each aliquot. The enzyme activity of IlvD in the cell extracts was immediately measured. The relative enzyme activity of IlvD in each cell extracts was plotted as a function of the NO exposure time (circles). IlvD remained fully active in the control E. coli cells after exposure to argon for 30 min (triangle). C), EPR spectra of recombinant IlvD. Spectrum a), IlvD purified from the E. coli cells before the NO exposure. Spectrum b), IlvD purified from the E. coli cells after 10 min NO exposure. Spectrum c), purified IlvD directly exposed to NO under anaerobic conditions. The protein concentration in each sample was about 3 μM. The data are the representatives of at least three independent experiments.
It has been shown that iron-sulfur proteins can be readily modified forming the protein-bound dinitrosyl iron complexes (DNICs) by NO in vitro and in vivo (Cruz-Ramos et al., 2002; Ding and Demple, 2000; Drapier, 1997; Foster and Cowan, 1999; Kennedy et al., 1997; Rogers et al., 2003). To examine whether the IlvD [4Fe-4S] cluster was also modified forming the IlvD-bound DNIC by the NO exposure, we purified recombinant IlvD from E. coli cells before and after the NO exposure. The unique EPR signal at g = 2.04 (Figure 2C) indicated that the IlvD [4Fe-4S] cluster was indeed converted to the IlvD-bound DNIC in vivo by the NO exposure. The EPR signal of the IlvD-bound DNIC purified from the NO-exposed E. coli cells was essentially identical to that of the purified IlvD [4Fe-4S] cluster directly exposed to NO in vitro (Figure 2C), suggesting that formation of the IlvD-bound DNIC in E. coli cells by NO does not require any additional factors. As the NO exposure time of E. coli cells was gradually increased, we found that the inactivation of IlvD in E. coli cells was inversely correlated with the formation of the IlvD-bound DNIC (data not shown). Collectively, these results suggest that NO inactivates IlvD by converting the IlvD [4Fe-4S] cluster to the IlvD-bound DNIC in E. coli cells.
Formation of the protein-bound DNICs in E. coli cells by NO
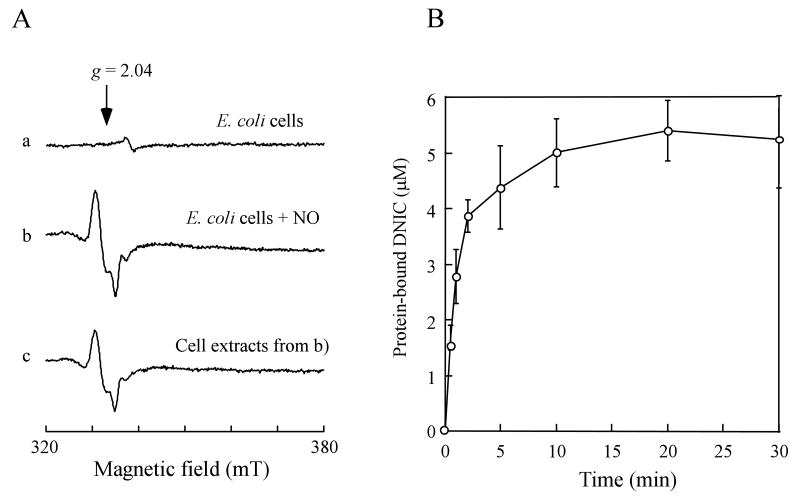
There are at least 200 unique iron-sulfur proteins in E. coli (Johnson et al., 2005). It is conceivable that other iron-sulfur proteins in cells may also be modified forming the protein-bound DNICs by the NO exposure. Figure 3A shows the EPR spectra of the wild-type E. coli cells (without recombinant IlvD) before and after the NO exposure under anaerobic conditions. The EPR signal at g = 2.04 indicated the formation of the protein-bound DNICs in E. coli cells by the NO exposure. The amplitude of the EPR signal at g = 2.04 reached the maximum when E. coli cells were exposed to NO for about 5 min under anaerobic conditions (Figure 3B). Further exposure of NO did not significantly increase the amplitude of the EPR signal at g = 2.04, indicating that modification of iron-sulfur proteins in E. coli cells by NO was nearly complete after 5 min NO exposure.
Figure 3. Formation of the protein-bound DNICs in E. coli cells by NO.
A), formation of the protein-bound DNICs in E. coli cells by NO. E. coli cells were exposed to pure NO gas at a rate of 100 nM per second for 0 min (spectrum a) and 10 min (spectrum b) using the Silastic tubing NO delivery system under anaerobic conditions. The E. coli cells were concentrated to O. D. at 600 nm of 10.0 after the NO exposure. Spectrum c), the cell extracts prepared from the NO-exposed E. coli cells. B), kinetics of the protein-bound DNICs formation in E. coli cells by NO. The amplitudes of the EPR signal at g = 2.04 in E. coli cells were plotted as a function of the NO exposure time. The endonuclease III-bound DNIC was used as a standard. The data in B) were the averages from three independent experiments.
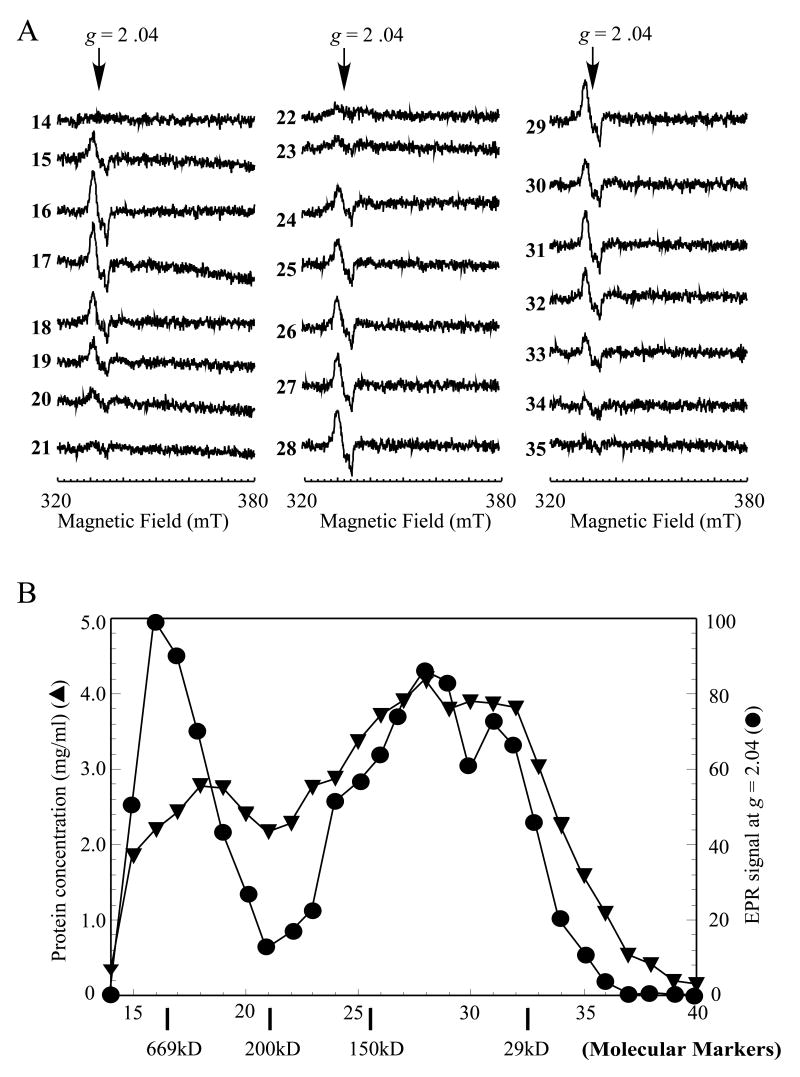
The cell extracts were then prepared from the NO-exposed E. coli cells and subjected to the EPR measurements. Over 80% of the EPR signal at g = 2.04 remained after the NO-exposed E. coli cells were disrupted by passing through French press (Figure 3A), supporting the idea that the protein-bound DNICs are stable in the cell extracts (Rogers and Ding, 2001). The cell extracts were further fractionated using a gel filtration column (Superdex-200) (Amersham Biosciences) as described in the Experimental Procedures. The EPR measurements of the eluted fractions showed that the protein-bound DNICs were present in almost all protein fractions (Figure 4A). Whereas the ratio of the protein-bound DNICs to the protein concentration in each fraction varied considerably (Figure 4B), the total amount of the protein-bound DNICs in the cell extracts was fully recovered in the eluted fractions. Thus, in addition to the IlvD-bound DNIC, a large number of different protein-bound DNICs were generated in E. coli cells by the NO exposure.
Figure 4. A broad distribution of the protein-bound DNICs in the NO-exposed E. coli cells.
The cell extracts were prepared from the NO-exposed E. coli cells as described in the Experimental Procedures. The cell extracts (20 mg total protein in 0.5 ml) were loaded onto a gel filtration column (superdex-200), and eluted at a flow rate of 0.5 ml per min using buffer containing Tris (20 mM, pH 8.0) and NaCl (500 mM). A), EPR spectra of the fractions eluted from the gel filtration column. B), the relative amplitude of the EPR signal at g = 2.04 (closed circles) and the protein concentration (mg/mL) (closed triangles) in each eluted fraction were plotted as a function of the fraction number. The molecular weights of the gel filtration protein standards are indicated on the bottom of x-axis.
The protein-bound DNICs are efficiently repaired in E. coli cells under aerobic growth conditions but not under anaerobic growth conditions
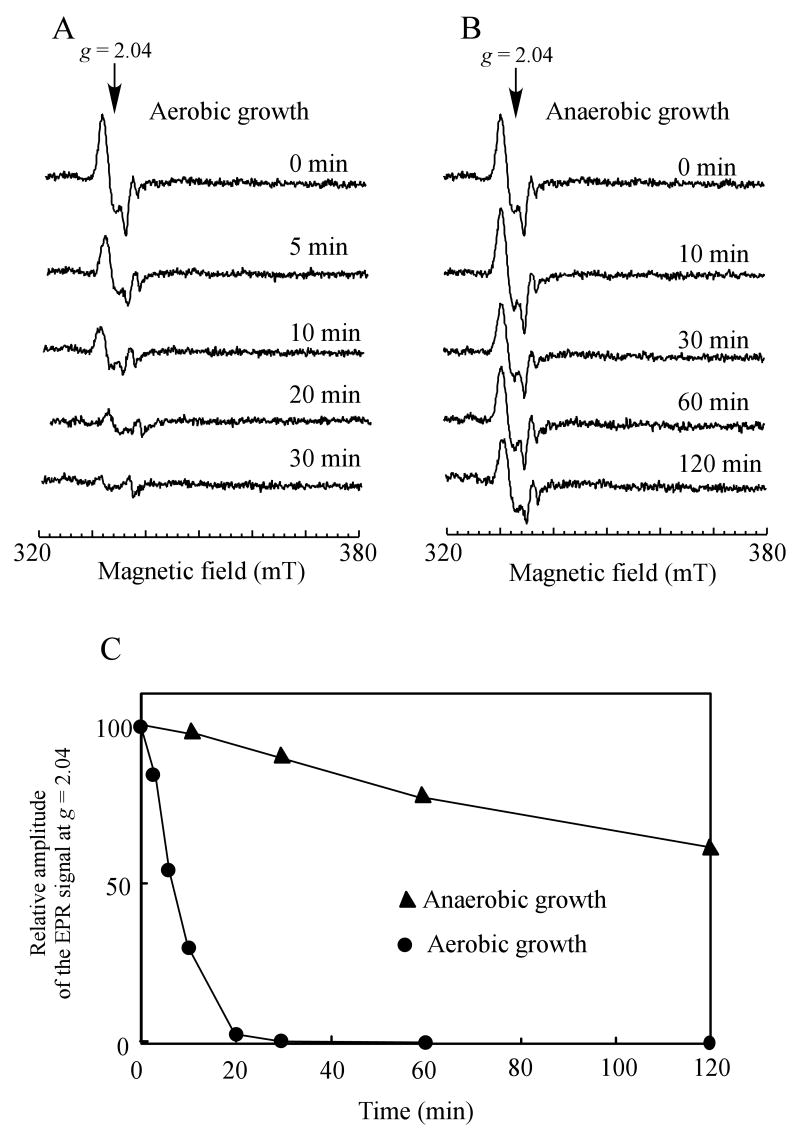
A broad distribution of the protein-bound DNICs in the NO-exposed E. coli cells suggests that multiple cellular functions could have been inactivated by the NO exposure. If cells are to survive, the protein-bound DNICs must be promptly repaired. Figure 5 shows that when the NO-exposed E. coli cells were returned to aerobic growth conditions, the EPR signal at g = 2.04 of the protein-bound DNICs quickly disappeared with a half-life time of about 5 min. The efficient repair of the protein-bound DNICs in aerobically growing E. coli cells did not require new protein synthesis, as addition of the protein synthesis inhibitor chloramphenicol had little or no effect on decay kinetics of the protein-bound DNICs in aerobically growing E. coli cells (data not shown). In contrast, when the NO-exposed E. coli cells were returned to anaerobic growth conditions, at least 75% of the protein-bound DNICs in E. coli cells remained after two hours incubation, indicating that anaerobically growing E. coli cells fail to repair the protein-bound DNICs.
Figure 5. Aerobically growing E. coli cells have a robust repair activity for the protein-bound DNICs.
Exponentially growing E. coli cells (concentrated to O.D. at 600 nm of 5.0) were exposed to pure NO gas at a rate of 100 nM per second for 10 min using the Silastic tubing NO delivery system anaerobically, followed by purge with pure argon gas. The NO-exposed cells were then returned to either aerobic or anaerobic growth conditions in minimal medium containing 0.2% glucose. A), the EPR spectra of the NO-exposed E. coli cells at indicated time after returned to aerobic growth conditions. B), the EPR spectra of the NO-exposed E. coli cells at indicated time after returned to anaerobic growth conditions. C), decay kinetics of the EPR signal at g = 2.04 of the NO-exposed E. coli cells under aerobic (closed circles) and anaerobic (closed triangles) growth conditions.
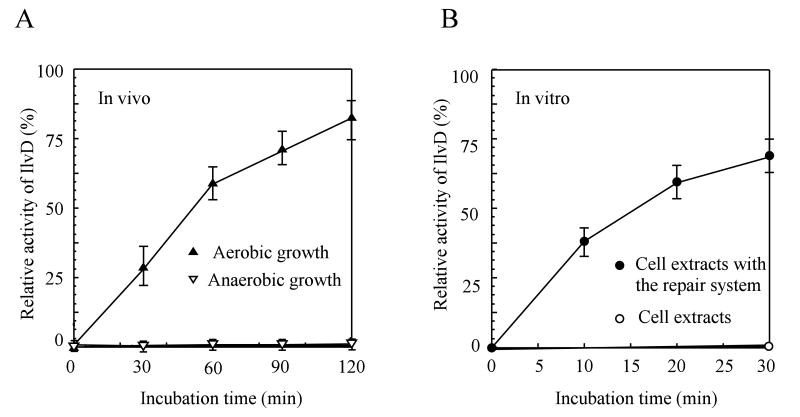
The NO-inactivated IlvD is quickly re-activated in E. coli cells under aerobic growth conditions but not under anaerobic growth conditions
To further examine whether the NO-modified iron-sulfur proteins are repaired in E. coli cells under aerobic growth conditions but not under anaerobic growth conditions, we measured the enzyme activity of IlvD in the cell extracts as described previously. Figure 6A shows that when the NO-exposed E. coli cells were returned to aerobic growth conditions, the NO-inactivated IlvD was quickly re-activated even in the presence of the protein synthesis inhibitor chloramphenicol. On the other hand, when the NO-exposed E. coli cells were returned to anaerobic growth conditions, the NO-inactivated IlvD remained inactive for at least two hours. Combining the results from the EPR spectra (Figure 5) and the enzyme activity measurements of IlvD in the cell extracts (Figure 6A), we concluded that E. coli cells can efficiently repair the NO-modified iron-sulfur proteins under aerobic growth conditions but not under anaerobic growth conditions, and that such a robust repair activity does not require new protein synthesis.
Figure 6. Re-activation of the NO-inactivated IlvD in E. coli cells under aerobic and anaerobic growth conditions.
Recombinant IlvD was expressed to about 2% of total cellular protein in E. coli cells before being subjected to the NO exposure for 10 min. A), re-activation of the NO-inactivated IlvD in E. coli cells under aerobic and anaerobic growth conditions. The enzyme activity of IlvD was measured in the cell extracts prepared from the NO-exposed E. coli cells after different incubation time under aerobic (filled triangles) or anaerobic (open triangles) growth conditions. Chloramphenicol (34 μg/mL) was added to the cell culture before the NO exposure to block any new protein synthesis. B), re-activation of the NO-inactivated IlvD in the cell extracts by re-assembly of iron-sulfur clusters. The cell extracts prepared from the NO-exposed E. coli cells containing recombinant IlvD were incubated with L-cysteine (0.5 mM), cysteine desulfurase IscS (1 μM), and Fe(NH4)2(SO4)2 (100 μM) in the presence of dithiothreitol (2 mM) anaerobically. Samples were taken at different incubation time. Closed circles: the enzyme activity of IlvD in the cell extracts after incubation with the iron-sulfur cluster repair system. Open circles: the enzyme activity of IlvD in the cell extracts without any additions. The data are the averages from three independent experiments.
Since the NO-inactivated IlvD was efficiently repaired in aerobically growing E. coli cells without new protein synthesis (Figure 6A), we postulated that re-assembly of iron-sulfur clusters may be sufficient for restoring the enzyme activity of the NO-inactivated IlvD in the cell extracts. In the experiments, the cell extracts prepared from the NO-exposed E. coli cells were incubated with L-cysteine, cysteine desulfurase IscS and ferrous iron in the presence of dithiothreitol at 37°C anaerobically, a system that was previously used for repairing the NO-modified iron-sulfur proteins (Rogers et al., 2003; Yang et al., 2002). As shown in Figure 6B, the NO-inactivated IlvD in the cell extracts was quickly re-activated after incubation with the iron-sulfur cluster repair system. About 70% of the IlvD enzyme activity in the NO-exposed cell extracts (relative to the untreated E. coli cells) was recovered after 30 min incubation. Missing any one of the components in the iron-sulfur cluster repair system failed to restore the enzyme activity of the NO-modified IlvD in the cell extracts (data not shown). The results demonstrated that reassembly of iron-sulfur clusters in the NO-modified IlvD is necessary and sufficient for reactivation of the NO-inactivated IlvD in the NO-exposed E. coli cells.
Potential role of L-cysteine in repairing the protein-bound DNICs in E. coli
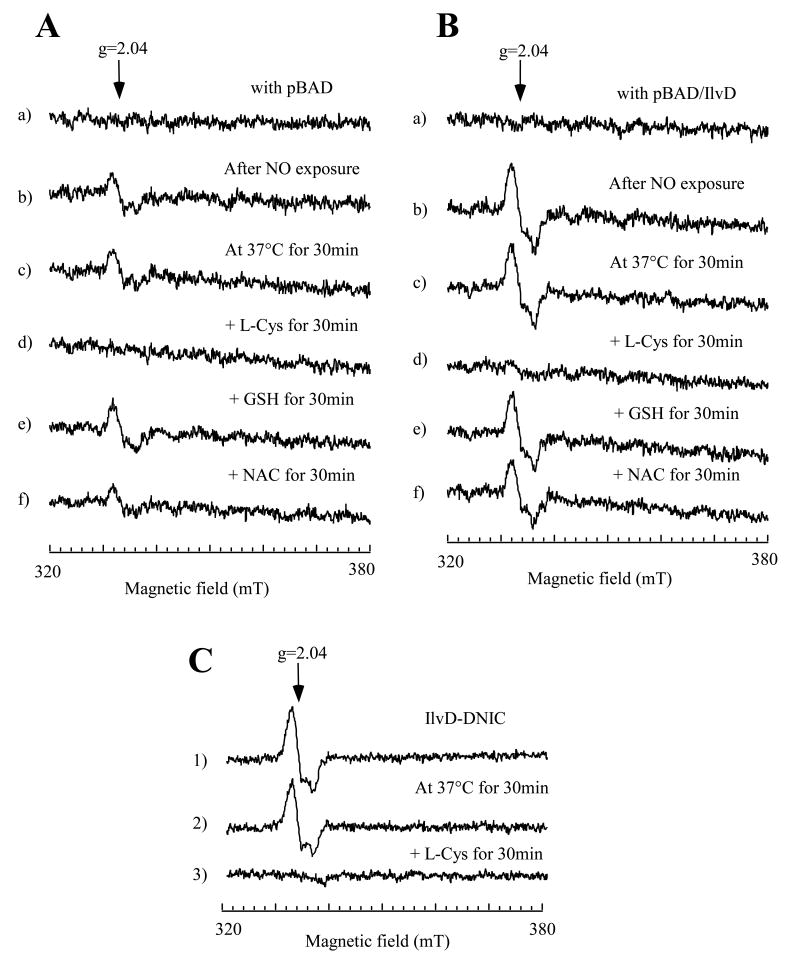
Because aerobically growing E. coli cells can efficiently repair the NO-inactivated IlvD without new protein synthesis (Figure 5 and Figure 6A), we reasoned that intracellular small molecules, but not specific proteins, may be directly responsible for the robust repair activity for the NO-modified iron-sulfur proteins observed in aerobically growing E. coli cells. Previous studies have shown that L-cysteine has its unique activity in decomposing the ferredoxin-bound DNIC in vitro (Rogers and Ding, 2001), and is also the substrate for cysteine desulfurase IscS (Zheng et al., 1998) to provide sulfide for the re-assembly of iron-sulfur clusters in the NO-modified proteins (Figure 6B) (Rogers et al., 2003; Yang et al., 2002). Conceivably, L-cysteine may have an important role in repairing the NO-modified proteins in E. coli cells.
To test the repair activity of L-cysteine in decomposing the IlvD-bound DNIC, we prepared the cell extracts from the E. coli cells containing either an expression vector or recombinant IlvD before and after the NO exposure. As shown in Figure 7A, the EPR signal at g = 2.04 of the cell extracts prepared from the NO-exposed E. coli cells without recombinant IlvD was completely eliminated after incubation with L-cysteine, but not with reduced glutathione or N-acetyl-L-cysteine. When the E. coli cells containing recombinant IlvD were subjected to the same NO exposure, the amplitude of the EPR signal at g = 2.04 was about 3-fold higher than that of the cells without recombinant IlvD (Figure 7B), further demonstrating the formation of the IlvD-bound DNIC in E. coli cells by NO. Again, the IlvD-bound DNIC in the cell extracts was efficiently decomposed by L-cysteine, but not by reduced glutathione or N-acetyl-L-cysteine (Figure 7B). We also incubated the purified IlvD-bound DNIC with L-cysteine, and found that L-cysteine was equally effective in decomposing the IlvD-bound DNIC (Figure 7C). While the mechanism underlying the L-cysteine-mediated decomposition of the protein-bound DNICs is still not well understood, these results suggested that L-cysteine may directly contribute to the robust repair activity for the protein-bound DNICs in aerobically growing E. coli cells. The physiological role of intracellular L-cysteine in repairing the NO-modified iron-sulfur proteins are currently under investigation.
Figure 7. L-cysteine can decompose the protein-bound DNICs in the cell extracts.
The cell extracts were prepared from the NO-exposed E. coli cells (concentrated to O. D. at 600 nm of 2.0) with the expression vector only (A) or with recombinant IlvD (B). Recombinant IlvD was induced to about 2% of total cellular proteins in E. coli cells. Spectrum a), the cell extracts prepared from the E. coli cells before the NO exposure. Spectrum b), the cell extracts prepared from the E. coli cells after the NO exposure. The cell extracts prepared from the NO-exposed E. coli cells were incubated with no addition (c), or L-cysteine (L-Cys) (1 mM) (d), reduced glutathione (GSH) (1 mM) (e), or N-acetyl-L-cysteine (NAC) (1 mM) (f) at 37°C for 30 min. C), purified IlvD-bound DNIC (5 μM) (1) was incubated with no addition (2) or L-cysteine (L-Cys) (1 mM) at 37°C for 30min. The samples were immediately transferred to EPR tubes after incubation. The EPR spectra were taken as described in the Experimental Procedures.
Discussion
In this study, we report that transient NO exposure effectively inhibits cell growth of E. coli in minimal medium under anaerobic growth conditions, and that cell growth is restored when the NO-exposed E. coli cells are either supplemented with the branched-chain amino acids (BCAA) anaerobically or returned to aerobic growth conditions. The enzyme activity measurements reveal that dihydroxyacid dehydratase (IlvD), an iron-sulfur enzyme essential for the BCAA biosynthesis in E. coli (Flint et al., 1993), is completely inactivated by the NO exposure with the concomitant formation of the IlvD-bound dinitrosyl iron complex (DNIC). Furthermore, we find that the NO-inactivated IlvD remains inactive in E. coli cells under anaerobic growth conditions, but is quickly re-activated under aerobic growth conditions even without new protein synthesis. The results not only substantiate the notion that IlvD is a sensitive target of the NO cytotoxicity (Hyduke et al., 2007), but also demonstrate that NO can effectively inhibit cell growth of E. coli under anaerobic growth conditions under which the NO-modified IlvD cannot be efficiently repaired.
Unlike reversible binding of NO to hemes in proteins (Ramachandran et al., 2002), the NO-mediated modification of iron-sulfur clusters is irreversible, associated with formation of the protein-bound DNIC (Cruz-Ramos et al., 2002; Ding and Demple, 2000; Drapier, 1997; Foster and Cowan, 1999; Kennedy et al., 1997; Rogers et al., 2003). Re-activation of the NO-modified iron-sulfur proteins requires decomposition of the DNIC in proteins, followed by re-assembly of new iron-sulfur clusters (Rogers et al., 2003). A number of proteins including cysteine desulfurase IscS (Yang et al., 2002) and an diiron protein YtfE (Justino et al., 2007) have been identified as critical for repairing the NO-modified iron-sulfur proteins. Both YtfE and IscS are highly induced in E. coli cells by NO exposure (Hyduke et al., 2007; Justino et al., 2005; Mukhopadhyay et al., 2004; Pullan et al., 2007). It is appealing to suggest that increased amounts of repair proteins such as IscS and YtfE may contribute to the robust repair activity for the NO-modified iron-sulfur proteins in aerobically growing E. coli cells. However, the efficient repair for the NO-modified iron-sulfur proteins in aerobically growing E. coli cells does not require new protein synthesis (Figure 5 and Figure 6A), suggesting that amounts of the specific repair enzymes are not the critical factors in determining the cellular repair activity for the NO-modified iron-sulfur proteins. In this context, we postulate that the amounts of intracellular small molecule(s) may be responsible for the robust repair reactivity in aerobically growing E. coli cells. We envision that the putative small molecules (e.g. ATP, L-cysteine, NADPH/NADH) are amply produced for repairing the NO-modified iron-sulfur proteins in E. coli cells under aerobic growth conditions, but not under anaerobic growth conditions. Because L-cysteine can decompose the protein-bound DNICs (Rogers and Ding, 2001) (Figure 7) and act as substrate for cysteine desulfurase IscS to provide sulfide for the iron-sulfur cluster re-assembly in proteins (Yang et al., 2002) (Figure 6B), we propose that L-cysteine may directly contribute to the robust repair activity for the NO-modified iron-sulfur proteins in aerobically growing E. coli cells. Nevertheless, it could not be excluded that other small molecules may also have their crucial roles in the cellular repair activity for the protein-bound DNICs in cells. Experiments are in progress to explore the physiological function of L-cysteine in repairing the NO-modified iron-sulfur proteins in E. coli cells.
Iron-sulfur proteins are involved in diverse physiological processes with over 200 unique iron-sulfur proteins identified so far (Johnson et al., 2005; Lill and Muhlenhoff, 2006). The gel filtration fractionation of the cell extracts prepared from the NO-exposed E. coli cells (Figure 4) clearly demonstrates that a large number of different iron-sulfur proteins are modified forming the protein-bound DNICs in cells by NO. The results provide additional evidence that iron-sulfur proteins are the primary targets of the NO cytotoxicity (Spiro, 2007). It should be pointed out that under defined experimental conditions, some iron-sulfur proteins are not essential. For example, because the aconitase [4Fe-4S] cluster (Gardner et al., 1997) of the citrate acid cycle is completely dispensable for anaerobic growth of E. coli, modification of the aconitase [4Fe-4S] cluster by NO would have no phenotype under anaerobic growth conditions. In contrast, because BCAA are required for cell growth of E. coli in minimal medium under aerobic or anaerobic growth conditions, inactivation of IlvD by NO would create the BCAA auxotrophy in E. coli. However, since the NO-inactivated IlvD is efficiently repaired in E. coli cells under aerobic growth conditions (Figure 6A), the NO exposure only induces a transitory depletion of BCAA and a brief delay of cell growth (Hyduke et al., 2007) (Figure 1). In contrast, the NO-inactivated IlvD remains inactive in E. coli cells under anaerobic growth conditions due to the deficient repair activity for the NO-inactivated IlvD (Figure 6A), and no BCAA are synthesized to support cell growth. A proposed model for the NO-induced bacteriostasis of E. coli under aerobic and anaerobic conditions is summarized in the Supplemental Figure 1.
Experimental Procedures
Cell growth and NO exposure
Overnight wild-type E. coli cells (GC4468) were diluted 1:100 in freshly prepared Luria-Bertani (LB) medium and incubated at 37°C with aeration (250 rpm) for 2.5 hours. For some experiments, overnight E. coli cells were grown in freshly prepared minimal medium containing 0.2% glucose. E. coli cells prepared from LB medium or minimal medium had similar response to the NO exposure in general under the experimental conditions. After harvested and washed once with minimal medium, cells were re-suspended in minimal medium containing 0.2% glucose. For the NO exposure, the Silastic tubing NO delivery system was used according to (Tamir et al., 1993). Briefly, pure NO gas (Air Co) was first passed through a soda-lima column to remove NO2 and higher oxides of nitrogen before being connected to the Silastic tubing. The length of the Silastic tubing (I.D. × O.D.; 0.025 × 0.047 in.) immersed in the cell culture was adjusted in such that about 100 nM NO per second was released to the cell culture in a sealed flask. The Silastic tubing NO delivery system provided a reproducible NO exposure without significantly changing pH of the cell culture. The chosen NO release rate was comparable to the reported NO production in activated polymorphonuclear leukocytes (Krieglstein et al., 2001) or in RAW 264.7 macrophages co-cultured with arginase-deficient Helicobacter pylori (Gobert et al., 2001). Typically, exponentially growing E. coli cells were purged with pure argon gas before being subjected to NO exposure. After the NO exposure, residual NO gas in cell cultures was re-purged with pure argon gas. The NO-exposed E. coli cells were then divided into two flasks: one was returned to anaerobic growth conditions, and the other to aerobic growth conditions. Each flask (1L) contained 100 mL cell culture was incubated at 37°C with shaking of 250 rpm. When indicated, amino acids (at final concentrations of 100 μg/L) were added to the cell cultures anaerobically. E. coli cells not treated with NO were used as controls. The cell growth was monitored in a Klett-Summerson photoelectric colorimeter (Klett MFG. Co.).
Cloning of E. coli dihydroxyacid dehydratase (ilvD)
The DNA fragment encoding gene ilvD was amplified from wild-type E. coli genomic DNA using the Polymerase Chain Reaction (PCR). Two primers (IlvD-1, 5′-gacactgctagcaaataaagtatgcctaag-3′; IlvD-2 5′-ggttgcggctcagccattattaacccccca-3′) were synthesized to contain the NheI site in one primer and the BlpI site in the other. The PCR product was digested with restriction enzymes NheI and BlpI, and the digested product was ligated to an expression vector pET28b+ to produce pTIlvD. Recombinant IlvD was expressed to about 2% of total cellular protein in E. coli cells using isopropyl β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 100 μM. In some experiments, recombinant IlvD was purified from the E. coli cell extracts using a Ni-agarose column followed by a HiTrap de-salting column (Amersham Biosciences) as described previously for other proteins (Rogers and Ding, 2001).
Enzyme activity assay of dihydroxyacid dehydratase (IlvD)
The cell extracts were prepared by passing the cells containing recombinant IlvD through French press once, followed by centrifugation at 34,000 g for 45 min. The specific enzyme activity was measured using substrate DL-2,3-dihydroxy-isovalerate which was synthesized according to the method of Cioffi et al. (Cioffi et al., 1980). All chemical reagents used in the DL-2,3-dihydroxy-isovalerate synthesis were obtained from Sigma-Aldrich (St. Louis, MO). In the enzyme assay, 10 μL cell extracts (∼5.0 mg total protein/mL) prepared from E. coli cells containing recombinant IlvD were added to 390 μL pre-incubated solutions containing 50 mM Tris (PH 8.0), 10 mM MgCl2, and 10 mM DL-2,3-dihydroxy-isovalerate (Flint et al., 1993). The reaction product (keto acids) was monitored at 240 nm using an extinction coefficient of 0.19 cm-1mM-1 (Flint et al., 1993).
Re-assembly of iron-sulfur clusters in the NO-inactivated IlvD
The cell extracts prepared from the NO-exposed E. coli cells containing recombinant IlvD (∼5.0 mg total protein/mL) were incubated with L-cysteine (0.5 mM), cysteine desulfurase IscS (2 μM), Fe(NH4)2(SO4)2 (100 μM), and dithiothreitol (2 mM) anaerobically at 37°C as described previously (Rogers et al., 2003; Yang et al., 2002). At different time points, aliquots (10 μL) of the reaction solution were taken for the IlvD enzyme activity assay as described above.
Fractionation of the cell extracts
The cell extracts were prepared from the NO-exposed E. coli cells by passing the cells through French press once, followed by centrifugation at 34,000 g for 45 min. The cell extracts were concentrated to about 40 mg protein/mL using the micro-concentrators (Millipore co.) and loaded onto a gel filtration column (Superdex-200) attached to a Fast Protein Liquid Chromatography (FPLC) system (Amersham Biosciences). The proteins were eluted from the column at a flow rate of 0.5 mL/min using a buffer containing Tris (20 mM, pH 8.0) and NaCl (500 mM). The gel filtration column was calibrated using a gel filtration protein standard (Sigma co.) under the same experimental conditions. The protein concentration in each eluted fraction was determined by the Bradford assay (Bio-Rad co).
EPR measurements of the protein-bound DNICs
The X-band EPR spectra were recorded using a Bruker model ESR-300 EPR spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. Routine EPR measurement conditions were: microwave frequency, 9.50 GHz; microwave power, 2.0 mW; modulation frequency, 100 kHz; modulation amplitude, 1.2 mT; sample temperature, 20 K; receive gain, 105. For quantification of the protein-bound DNICs, the E. coli endonuclease III-bound DNIC (Rogers et al., 2003) was used as a standard. The concentration of the endonuclease III-bound DNICs was pre-determined as described previously (Rogers et al., 2003).
Supplementary Material
Acknowledgments
We are grateful to Dr. James C. Liao (UCLA) for his kind advice on synthesizing DL-2,3-dihydroxy-isovalerate. This work was supported by Public Health Service Grant (RO1 CA107494) from the National Institutes of Health to H.D.
Abbreviations
- BCAA
the branched-chain amino acids
- DNIC
dinitrosyl iron complex
- EPR
electron paramagnetic resonance
- IlvD
dihydroxyacid dehydratase
- LeuCD
isopropylmalate isomerase
References
- Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi EA, Shaw KJ, Bailey WF, Berg CM. Improved synthesis of the sodium salt of DL-alpha, beta-dihydroxyisovaleric acid. Anal Biochem. 1980;104:485–488. doi: 10.1016/0003-2697(80)90104-9. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. Embo J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Autreaux B, Tucker NP, Dixon R, Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci U S A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier JC. Interplay between NO and [Fe-S] clusters: relevance to biological systems. Methods. 1997;11:319–329. doi: 10.1006/meth.1996.0426. [DOI] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Finnegan MG, Fu W, Johnson MK. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J Biol Chem. 1993;268:14732–14742. [PubMed] [Google Scholar]
- Foster MW, Cowan JA. Chemistry of Nitric Oxide with Protein-Bound Iron Sulfur Centers. Insights on Physiological Reactivity. J Am Chem Soc. 1999;121:4093–4100. [Google Scholar]
- Gardner AM, Gardner PR. Flavohemoglobin Detoxifies Nitric Oxide in Aerobic, but Not Anaerobic, Escherichia coli. EVIDENCE FOR A NOVEL INDUCIBLE ANAEROBIC NITRIC OXIDE- SCAVENGING ACTIVITY. J Biol Chem. 2002;277:8166–8171. doi: 10.1074/jbc.M110470200. [DOI] [PubMed] [Google Scholar]
- Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an Inducible Catalyst for Nitric Oxide Reduction and Detoxification in Escherichia coli. J Biol Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- Gardner PR, Costantino G, Szabo C, Salzman AL. Nitric oxide sensitivity of the aconitases. J Biol Chem. 1997;272:25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
- Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CM, Giuffre A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J Biol Chem. 2002;277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignarro LJ. Nitric oxide: a unique endogenous signaling molecule in vascular biology. Biosci Rep. 1999;19:51–71. doi: 10.1023/a:1020150124721. [DOI] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Justino MC, Vicente JB, Teixeira M, Saraiva LM. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- Justino MC, Almeida CC, Teixeira M, Saraiva LM. Escherichia coli Di-iron YtfE Protein Is Necessary for the Repair of Stress-damaged Iron-Sulfur Clusters. J Biol Chem. 2007;282:10352–10359. doi: 10.1074/jbc.M610656200. [DOI] [PubMed] [Google Scholar]
- Kennedy MC, Antholine WE, Beinert H. An EPR investigation of the products of the reaction of cytosolic and mitochondrial aconitases with nitric oxide. J Biol Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- Krieglstein CF, Cerwinka WH, Laroux FS, Salter JW, Russell JM, Schuermann G, Grisham MB, Ross CR, Granger DN. Regulation of murine intestinal inflammation by reactive metabolites of oxygen and nitrogen: divergent roles of superoxide and nitric oxide. J Exp Med. 2001;194:1207–1218. doi: 10.1084/jem.194.9.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poock SR, Leach ER, Moir JW, Cole JA, Richardson DJ. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J Biol Chem. 2002;277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- Poole RK, Hughes MN. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 2000;36:775–783. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TM, Read RC, Green J, Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Levonen AL, Brookes PS, Ceaser E, Shiva S, Barone MC, Darley-Usmar V. Mitochondria, nitric oxide, and cardiovascular dysfunction. Free Radic Biol Med. 2002;33:1465–1474. doi: 10.1016/s0891-5849(02)01142-5. [DOI] [PubMed] [Google Scholar]
- Rogers PA, Ding H. L-cysteine-mediated destabilization of dinitrosyl iron complexes in proteins. J Biol Chem. 2001;276:30980–30986. doi: 10.1074/jbc.M101037200. [DOI] [PubMed] [Google Scholar]
- Rogers PA, Eide L, Klungland A, Ding H. Reversible inactivation of E. coli endonuclease III by nitric oxide via modification of its [4Fe-4S] cluster. DNA Repair. 2003;2:809–817. doi: 10.1016/s1568-7864(03)00065-x. [DOI] [PubMed] [Google Scholar]
- Spiro S. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- Tamir S, Lewis RS, de Rojas Walker T, Deen WM, Wishnok JS, Tannenbaum SR. The influence of delivery rate on the chemistry and biological effects of nitric oxide. Chem Res Toxicol. 1993;6:895–899. doi: 10.1021/tx00036a021. [DOI] [PubMed] [Google Scholar]
- Wallace MA, Liou LL, Martins J, Clement MH, Bailey S, Longo VD, Valentine JS, Gralla EB. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J Biol Chem. 2004;279:32055–32062. doi: 10.1074/jbc.M403590200. [DOI] [PubMed] [Google Scholar]
- Woodmansee AN, Imlay JA. A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol Microbiol. 2003;49:11–22. doi: 10.1046/j.1365-2958.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Rogers PA, Ding H. Repair of nitric oxide modified ferredoxin [2Fe-2S] cluster by cysteine desulfurase (IscS) J Biol Chem. 2002;277:12868–12873. doi: 10.1074/jbc.M109485200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.