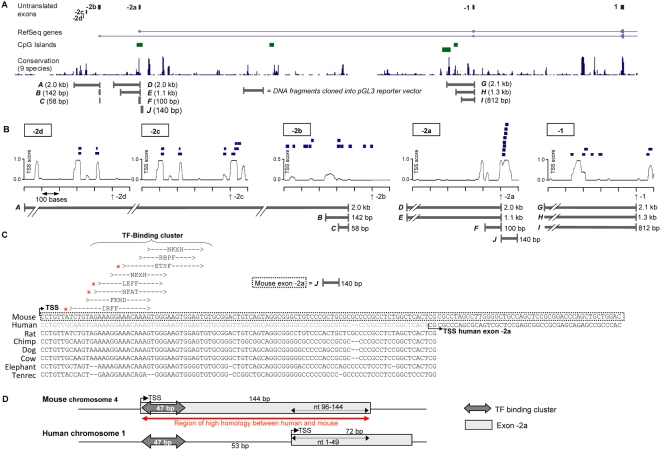
Figure 7. Bioinformatic analysis of potential promoter elements and TF binding sites in PIK3CD.
(A) Schematic representation of the murine p110δ 5′UTR. The upper panel shows the five untranslated murine p110δ exons (and exon 1) in their genomic context, with below (in descending order): the mouse RefSeq genes, CpG islands, homology with 9 species (rat, human, chimp, rhesus, dog, cow, armadillo, elephant, tenrec, plotted against the murine sequence) and the genomic fragments that were subcloned for use in gene reporter assays. (B) Locations of the conserved TF binding sites in the 600 bases (500 upstream to 100 downstream of the transcript start site) in the forward strand flanking the different 5′untranslated exons of mouse p110δ gene. The exon start sites are indicated by the vertical arrows, the TF binding sites found on the forward strand are shown as blue boxes above the TSS score graphs. Also shown is the degree of cross species (28 species) genomic conservation as calculated by the phastCons program [94] from a minimum of 0.0 to a maximum of 1.0. The genomic DNA fragments subcloned into the PGL3 reporter vector are shown underneath. (C) Alignment and conservation of the TF binding cluster identified in mouse exon -2a with genomic sequences upstream of the translation start site of PIK3CD of 7 other species. (D) Schematic representation of TF binding cluster location in relation to exon -2 in human and mouse.